Inverse In Vitro Selection Enables Comprehensive Analysis of Cross-Chiral L-Aptamer Interactions
- PMID: 36282114
- PMCID: PMC9798143
- DOI: 10.1002/cbic.202200520
Inverse In Vitro Selection Enables Comprehensive Analysis of Cross-Chiral L-Aptamer Interactions
Abstract
Aptamers composed of mirror-image L-(deoxy)ribose nucleic acids, referred to as L-aptamers, are a promising class of RNA-binding reagents. Yet, the selectivity of cross-chiral interactions between L-aptamers and their RNA targets remain poorly characterized, limiting the potential utility of this approach for applications in biological systems. Herein, we carried out the first comprehensive analysis of cross-chiral L-aptamer selectivity using a newly developed "inverse" in vitro selection approach that exploits the genetic nature of the D-RNA ligand. By employing a library of more than a million target-derived sequences, we determined the RNA sequence and structural preference of a model L-aptamer and revealed previously unidentified and potentially broad off-target RNA binding behaviors. These results provide valuable information for assessing the likelihood and consequences of potential off-target interactions and reveal strategies to mitigate these effects. Thus, inverse in vitro selection provides several opportunities to advance L-aptamer technology.
Keywords: In vitro selection; L-aptamers; RNA; selectivity; sequencing.
© 2022 Wiley-VCH GmbH.
Figures

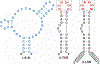
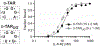
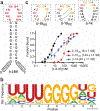
References
-
- Guan L, Disney MD, ACS Chem. Biol 2012, 7, 73–86; - PubMed
- Shortridge MD, Varani G, Curr. Opin. Struct. Biol 2015, 30, 79–88; - PMC - PubMed
- Thomas JR, Hergenrother PJ, Chem. Rev 2008, 108, 1171–1224; - PubMed
- Velagapudi SP, Gallo SM, Disney MD, Nat. Chem. Biol 2014, 10, 292–297; - PMC - PubMed
- Pal S, ‘t Hart P, Front. Mol. Biosci 2022, 9; - PMC - PubMed
- Chen Y, Yang F, Zubovic L, Pavelitz T, Yang W, Godin K, Walker M, Zheng S, Macchi P, Varani G, Nat. Chem. Biol 2016, 12, 717–723; - PMC - PubMed
- Koirala D, Lewicka A, Koldobskaya Y, Huang H, Piccirilli JA, ACS Chem. Biol 2020, 15, 205–216. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

