GlycoEnzOnto: a GlycoEnzyme pathway and molecular function ontology
- PMID: 36282863
- PMCID: PMC9750110
- DOI: 10.1093/bioinformatics/btac704
GlycoEnzOnto: a GlycoEnzyme pathway and molecular function ontology
Abstract
Motivation: The 'glycoEnzymes' include a set of proteins having related enzymatic, metabolic, transport, structural and cofactor functions. Currently, there is no established ontology to describe glycoEnzyme properties and to relate them to glycan biosynthesis pathways.
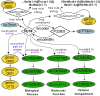
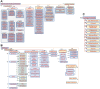
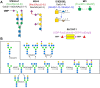
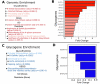
Results: We present GlycoEnzOnto, an ontology describing 403 human glycoEnzymes curated along 139 glycosylation pathways, 134 molecular functions and 22 cellular compartments. The pathways described regulate nucleotide-sugar metabolism, glycosyl-substrate/donor transport, glycan biosynthesis and degradation. The role of each enzyme in the glycosylation initiation, elongation/branching and capping/termination phases is described. IUPAC linear strings present systematic human/machine-readable descriptions of individual reaction steps and enable automated knowledge-based curation of biochemical networks. All GlycoEnzOnto knowledge is integrated with the Gene Ontology biological processes. GlycoEnzOnto enables improved transcript overrepresentation analyses and glycosylation pathway identification compared to other available schema, e.g. KEGG and Reactome. Overall, GlycoEnzOnto represents a holistic glycoinformatics resource for systems-level analyses.
Availability and implementation: https://github.com/neel-lab/GlycoEnzOnto.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2022. Published by Oxford University Press.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

