ATP13A1 prevents ERAD of folding-competent mislocalized and misoriented proteins
- PMID: 36283413
- PMCID: PMC9675726
- DOI: 10.1016/j.molcel.2022.09.035
ATP13A1 prevents ERAD of folding-competent mislocalized and misoriented proteins
Abstract
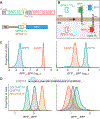
The biosynthesis of thousands of proteins requires targeting a signal sequence or transmembrane segment (TM) to the endoplasmic reticulum (ER). These hydrophobic ɑ helices must localize to the appropriate cellular membrane and integrate in the correct topology to maintain a high-fidelity proteome. Here, we show that the P5A-ATPase ATP13A1 prevents the accumulation of mislocalized and misoriented proteins, which are eliminated by different ER-associated degradation (ERAD) pathways in mammalian cells. Without ATP13A1, mitochondrial tail-anchored proteins mislocalize to the ER through the ER membrane protein complex and are cleaved by signal peptide peptidase for ERAD. ATP13A1 also facilitates the topogenesis of a subset of proteins with an N-terminal TM or signal sequence that should insert into the ER membrane with a cytosolic N terminus. Without ATP13A1, such proteins accumulate in the wrong orientation and are targeted for ERAD by distinct ubiquitin ligases. Thus, ATP13A1 prevents ERAD of diverse proteins capable of proper folding.
Keywords: ER-associated degradation; protein localization; protein topology; quality control; signal sequence; transmembrane proteins.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
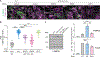
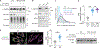


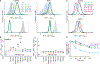
References
-
- Carvalho P, Goder V, and Rapoport TA (2006). Distinct Ubiquitin-Ligase Complexes Define Convergent Pathways for the Degradation of ER Proteins. Cell 126, 361–373. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

