A high-content endogenous GLUT4 trafficking assay reveals new aspects of adipocyte biology
- PMID: 36283703
- PMCID: PMC9595207
- DOI: 10.26508/lsa.202201585
A high-content endogenous GLUT4 trafficking assay reveals new aspects of adipocyte biology
Abstract
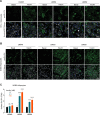
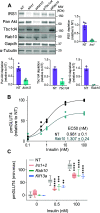
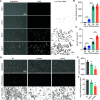
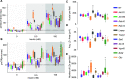
Insulin-induced GLUT4 translocation to the plasma membrane in muscle and adipocytes is crucial for whole-body glucose homeostasis. Currently, GLUT4 trafficking assays rely on overexpression of tagged GLUT4. Here we describe a high-content imaging platform for studying endogenous GLUT4 translocation in intact adipocytes. This method enables high fidelity analysis of GLUT4 responses to specific perturbations, multiplexing of other trafficking proteins and other features including lipid droplet morphology. Using this multiplexed approach we showed that Vps45 and Rab14 are selective regulators of GLUT4, but Trarg1, Stx6, Stx16, Tbc1d4 and Rab10 knockdown affected both GLUT4 and TfR translocation. Thus, GLUT4 and TfR translocation machinery likely have some overlap upon insulin-stimulation. In addition, we identified Kif13A, a Rab10 binding molecular motor, as a novel regulator of GLUT4 traffic. Finally, comparison of endogenous to overexpressed GLUT4 highlights that the endogenous GLUT4 methodology has an enhanced sensitivity to genetic perturbations and emphasises the advantage of studying endogenous protein trafficking for drug discovery and genetic analysis of insulin action in relevant cell types.
© 2022 Diaz-Vegas et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
