Translation Rescue by Targeting Ppp1r15a through Its Upstream Open Reading Frame in Sepsis-Induced Acute Kidney Injury in a Murine Model
- PMID: 36283811
- PMCID: PMC10103092
- DOI: 10.1681/ASN.2022060644
Translation Rescue by Targeting Ppp1r15a through Its Upstream Open Reading Frame in Sepsis-Induced Acute Kidney Injury in a Murine Model
Abstract
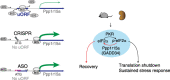
Background: Translation shutdown is a hallmark of late-phase, sepsis-induced kidney injury. Methods for controlling protein synthesis in the kidney are limited. Reversing translation shutdown requires dephosphorylation of the eukaryotic initiation factor 2 (eIF2) subunit eIF2 α ; this is mediated by a key regulatory molecule, protein phosphatase 1 regulatory subunit 15A (Ppp1r15a), also known as GADD34.
Methods: To study protein synthesis in the kidney in a murine endotoxemia model and investigate the feasibility of translation control in vivo by boosting the protein expression of Ppp1r15a, we combined multiple tools, including ribosome profiling (Ribo-seq), proteomics, polyribosome profiling, and antisense oligonucleotides, and a newly generated Ppp1r15a knock-in mouse model and multiple mutant cell lines.
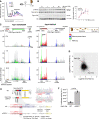
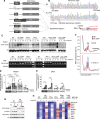
Results: We report that translation shutdown in established sepsis-induced kidney injury is brought about by excessive eIF2 α phosphorylation and sustained by blunted expression of the counter-regulatory phosphatase Ppp1r15a. We determined the blunted Ppp1r15a expression persists because of the presence of an upstream open reading frame (uORF). Overcoming this barrier with genetic and antisense oligonucleotide approaches enabled the overexpression of Ppp1r15a, which salvaged translation and improved kidney function in an endotoxemia model. Loss of this uORF also had broad effects on the composition and phosphorylation status of the immunopeptidome-peptides associated with the MHC-that extended beyond the eIF2 α axis.
Conclusions: We found Ppp1r15a is translationally repressed during late-phase sepsis because of the existence of an uORF, which is a prime therapeutic candidate for this strategic rescue of translation in late-phase sepsis. The ability to accurately control translation dynamics during sepsis may offer new paths for the development of therapies at codon-level precision.
Podcast: This article contains a podcast at.
Copyright © 2022 by the American Society of Nephrology.
Conflict of interest statement
E.H. Doud reports receiving honoraria from PEAKS Bioinformatics Solutions. A. Halim reports having ownership interest in OVIBIO Corporation. All remaining authors have nothing to disclose.
Figures




Comment in
-
Targeting a Single Codon to Rescue Septic Acute Kidney Injury.J Am Soc Nephrol. 2023 Feb 1;34(2):179-181. doi: 10.1681/ASN.0000000000000021. Epub 2023 Jan 17. J Am Soc Nephrol. 2023. PMID: 36735369 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

