An expanded lexicon for the ubiquitin code
- PMID: 36284179
- PMCID: PMC9595094
- DOI: 10.1038/s41580-022-00543-1
An expanded lexicon for the ubiquitin code
Abstract
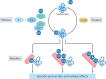
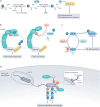
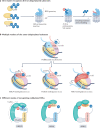
Our understanding of the ubiquitin code has greatly evolved from conventional E1, E2 and E3 enzymes that modify Lys residues on specific substrates with a single type of ubiquitin chain to more complex processes that regulate and mediate ubiquitylation. In this Review, we discuss recently discovered endogenous mechanisms and unprecedented pathways by which pathogens rewrite the ubiquitin code to promote infection. These processes include unconventional ubiquitin modifications involving ester linkages with proteins, lipids and sugars, or ubiquitylation through a phosphoribosyl bridge involving Arg42 of ubiquitin. We also introduce the enzymatic pathways that write and reverse these modifications, such as the papain-like proteases of severe acute respiratory syndrome coronavirus (SARS-CoV) and SARS-CoV-2. Furthermore, structural studies have revealed that the ultimate functions of ubiquitin are mediated not simply by straightforward recognition by ubiquitin-binding domains. Instead, elaborate multivalent interactions between ubiquitylated targets or ubiquitin chains and their readers (for example, the proteasome, the MLL1 complex or DOT1L) can elicit conformational changes that regulate protein degradation or transcription. The newly discovered mechanisms provide opportunities for innovative therapeutic interventions for diseases such as cancer and infectious diseases.
© 2022. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

