STtools: A Comprehensive Software Pipeline for Ultra-high Resolution Spatial Transcriptomics Data
- PMID: 36284674
- PMCID: PMC9590442
- DOI: 10.1093/bioadv/vbac061
STtools: A Comprehensive Software Pipeline for Ultra-high Resolution Spatial Transcriptomics Data
Abstract
Motivation: While there are many software pipelines for analyzing spatial transcriptomics data, few can process ultra high-resolution datasets generated by emerging technologies. There is a clear need for new software tools that can handle sub-micrometer resolution spatial transcriptomics data with computational scalability without compromising its resolution.
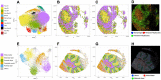
Results: We developed STtools, a software pipeline that provides a versatile framework to handle spatial transcriptomics datasets with various resolutions, such as the ones produced by Seq-Scope (<1μm), Slide-seq (10μm) and VISIUM (100μm). It automatically processes raw FASTQ files and runs downstream analyses at several folds higher resolution than existing methods. It also generates various visualizations including transcriptome density, cell type mapping, marker gene highlighting, and subcellular architectures.
Availability: STtools is publically available for download at https://github.com/seqscope/STtools.
Conflict of interest statement
Conflict of Interest: JX is an employee of Intel Corp. HMK is an employee of Regeneron Pharmaceuticals, he owns stock and stock options for Regeneron Pharmaceuticals. JHL is an inventor on pending patent applications related to Seq-Scope.
Figures
References
-
- 10x Genomics (2022) Space ranger: spatial transcriptomics. https://support.10xgenomics.com/spatial-gene-expression/software/pipelin... (1 May 2022, date last accessed).
-
- Kaminow B. et al. (2021) STARsolo: accurate, fast and versatile mapping/quantification of single-cell and single-nucleus RNA-seq data. bioRxiv doi:10.1101/2021.05.05.442755.
-
- Li H. (2021) Seqtk. https://github.com/lh3/seqtk (1 May 2022, date last accessed).
Grants and funding
LinkOut - more resources
Full Text Sources

