A Novel Protein Encoded by Exosomal CircATG4B Induces Oxaliplatin Resistance in Colorectal Cancer by Promoting Autophagy
- PMID: 36285810
- PMCID: PMC9762280
- DOI: 10.1002/advs.202204513
A Novel Protein Encoded by Exosomal CircATG4B Induces Oxaliplatin Resistance in Colorectal Cancer by Promoting Autophagy
Abstract
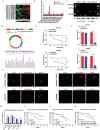
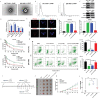
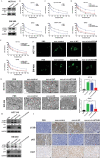
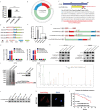
Oxaliplatin is commonly used in chemotherapeutic regimens for colorectal cancer (CRC) after surgical resection. However, acquired chemoresistance seriously affects the curative effect in CRC patients, and the mechanism is still unclear. Here, a circular RNA, circATG4B is identified, which plays an important role in oxaliplatin resistance in CRC. circATG4B expression is found to be increased in exosomes secreted by oxaliplatin-resistant CRC cells. In addition, the results suggest that circATG4B induces oxaliplatin resistance by promoting autophagy. Further in vivo and in vitro studies indicate that the effect of circATG4B is attributed to its potential to encode a novel protein, circATG4B-222aa. Next, circATG4B-222aa is found to function as a decoy to competitively interact with TMED10 and prevent TMED10 from binding to ATG4B, which leads to increased autophagy followed by induction of chemoresistance. Therefore, this study reveals that exosomal circATG4B participates in the decreased chemosensitivity of CRC cells, providing a new rationale for a potential therapeutic target for oxaliplatin resistance in CRC.
Keywords: autophagy; chemoresistance; circular RNA; colorectal cancer (CRC).
© 2022 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., Bray F., CA Cancer J. Clin. 2021, 71, 209. - PubMed
-
- Jardim D. L., Rodrigues C. A., Novis Y. A. S., Rocha V. G., Hoff P. M., Ann. Oncol. 2012, 23, 1937. - PubMed
-
- Théry C., Zitvogel L., Amigorena S., Nat. Rev. Immunol. 2002, 2, 569. - PubMed
-
- van Niel G., Porto‐Carreiro I., Simoes S., Raposo G., J. Biochem. 2006, 140, 13. - PubMed
-
- Kulcheski F. R., Christoff A. P., Margis R., J. Biotechnol. 2016, 238, 42. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 82002508/National Natural Science Foundation of China
- 82002928/National Natural Science Foundation of China
- 81900597/National Natural Science Foundation of China
- 82073171/National Natural Science Foundation of China
- 2021A1515012136/Natural Science Foundation of Guangdong Province
- 201904010020/Science and Technology Program Guangzhou
- 202102080038/Science and Technology Program Guangzhou
- 202201010898/Science and Technology Program Guangzhou
- 2022YW030009/National key Clinical Specialty Construction Project
- KY012021186/Excellent Young Talent Program of Guangdong Provincial People's Hospital
- KY012021159/GDPH supporting Funding for NSFC Program
LinkOut - more resources
Full Text Sources
Medical
