GUBS: Graph-Based Unsupervised Brain Segmentation in MRI Images
- PMID: 36286356
- PMCID: PMC9604689
- DOI: 10.3390/jimaging8100262
GUBS: Graph-Based Unsupervised Brain Segmentation in MRI Images
Abstract
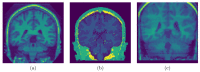
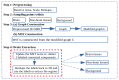
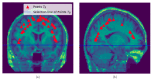
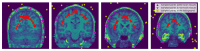
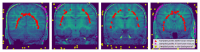
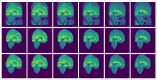
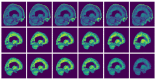
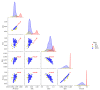
Brain segmentation in magnetic resonance imaging (MRI) images is the process of isolating the brain from non-brain tissues to simplify the further analysis, such as detecting pathology or calculating volumes. This paper proposes a Graph-based Unsupervised Brain Segmentation (GUBS) that processes 3D MRI images and segments them into brain, non-brain tissues, and backgrounds. GUBS first constructs an adjacency graph from a preprocessed MRI image, weights it by the difference between voxel intensities, and computes its minimum spanning tree (MST). It then uses domain knowledge about the different regions of MRIs to sample representative points from the brain, non-brain, and background regions of the MRI image. The adjacency graph nodes corresponding to sampled points in each region are identified and used as the terminal nodes for paths connecting the regions in the MST. GUBS then computes a subgraph of the MST by first removing the longest edge of the path connecting the terminal nodes in the brain and other regions, followed by removing the longest edge of the path connecting non-brain and background regions. This process results in three labeled, connected components, whose labels are used to segment the brain, non-brain tissues, and the background. GUBS was tested by segmenting 3D T1 weighted MRI images from three publicly available data sets. GUBS shows comparable results to the state-of-the-art methods in terms of performance. However, many competing methods rely on having labeled data available for training. Labeling is a time-intensive and costly process, and a big advantage of GUBS is that it does not require labels.
Keywords: brain tissues; minimum spanning tree; non-brain tissues; segmentation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Li J., Erdt M., Janoos F., Chang T.C., Egger J. Computer-Aided Oral and Maxillofacial Surgery. Academic Press; Cambridge, MA, USA: 2021. Medical image segmentation in oral-maxillofacial surgery; pp. 1–27.
Grants and funding
LinkOut - more resources
Full Text Sources

