Unraveling Capsule Biosynthesis and Signaling Networks in Cryptococcus neoformans
- PMID: 36287085
- PMCID: PMC9769619
- DOI: 10.1128/spectrum.02866-22
Unraveling Capsule Biosynthesis and Signaling Networks in Cryptococcus neoformans
Abstract
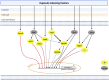
The polysaccharide capsule of Cryptococcus neoformans-an opportunistic basidiomycete pathogen and the major etiological agent of fungal meningoencephalitis-is a key virulence factor that prevents its phagocytosis by host innate immune cells. However, the complex signaling networks for their synthesis and attachment remain elusive. In this study, we systematically analyzed capsule biosynthesis and signaling networks using C. neoformans transcription factor (TF) and kinase mutant libraries under diverse capsule-inducing conditions. We found that deletion of GAT201, YAP1, BZP4, and ADA2 consistently caused capsule production defects in all tested media, indicating that they are capsule-regulating core TFs. Epistatic and expression analyses showed that Yap1 and Ada2 control Gat201 upstream, whereas Bzp4 and Gat201 independently regulate capsule production. Next, we searched for potential upstream kinases and found that mutants lacking PKA1, BUD32, POS5, IRE1, or CDC2801 showed reduced capsule production under all three capsule induction conditions, whereas mutants lacking HOG1 and IRK5 displayed enhanced capsule production. Pka1 and Irk5 controlled the induction of GAT201 and BZP4, respectively, under capsule induction conditions. Finally, we monitored the transcriptome profiles governed by Bzp4, Gat201, and Ada2 under capsule-inducing conditions and demonstrated that these TFs regulate redundant and unique sets of downstream target genes. Bzp4, Ada2, and Gat201 govern capsule formation in C. neoformans by regulating the expression of various capsule biosynthesis genes and chitin/chitosan synthesis genes in a positive and negative manner, respectively. In conclusion, this study provides further insights into the complex regulatory mechanisms of capsule production-related signaling pathways in C. neoformans. IMPORTANCE Over the past decades, human fungal pathogens, including C. neoformans, have emerged as a major public threat since the AIDS pandemic, only to gain more traction in connection to COVID-19. Polysaccharide capsules are rare fungal virulence factors that are critical for protecting C. neoformans from phagocytosis by macrophages. To date, more than 75 proteins involved in capsule synthesis and cell wall attachment have been reported in C. neoformans; however, their complex upstream signaling networks remain elusive. In this study, we demonstrated that Ada2, Yap1, Bzp4, and Gat201 were key capsule-inducing transcriptional regulators. Yap1 and Ada2 function upstream of Gat201, whereas Bzp4 and Gat201 function independently. Genome-wide transcriptome profiling revealed that Bzp4, Gat201, and Ada2 promote capsule production and attachment by positively and negatively regulating genes involved in capsule synthesis and chitin/chitosan synthesis, respectively. Thus, this study provides comprehensive insights into the complex capsule-regulating signaling pathway in C. neoformans.
Keywords: Ada2; Bzp4; Gat201; Yap1; kinase; transcription factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

