Functional antibodies exhibit light chain coherence
- PMID: 36289331
- PMCID: PMC9607724
- DOI: 10.1038/s41586-022-05371-z
Functional antibodies exhibit light chain coherence
Abstract
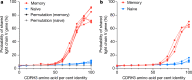
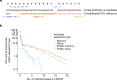
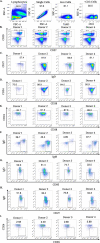
The vertebrate adaptive immune system modifies the genome of individual B cells to encode antibodies that bind particular antigens1. In most mammals, antibodies are composed of heavy and light chains that are generated sequentially by recombination of V, D (for heavy chains), J and C gene segments. Each chain contains three complementarity-determining regions (CDR1-CDR3), which contribute to antigen specificity. Certain heavy and light chains are preferred for particular antigens2-22. Here we consider pairs of B cells that share the same heavy chain V gene and CDRH3 amino acid sequence and were isolated from different donors, also known as public clonotypes23,24. We show that for naive antibodies (those not yet adapted to antigens), the probability that they use the same light chain V gene is around 10%, whereas for memory (functional) antibodies, it is around 80%, even if only one cell per clonotype is used. This property of functional antibodies is a phenomenon that we call light chain coherence. We also observe this phenomenon when similar heavy chains recur within a donor. Thus, although naive antibodies seem to recur by chance, the recurrence of functional antibodies reveals surprising constraint and determinism in the processes of V(D)J recombination and immune selection. For most functional antibodies, the heavy chain determines the light chain.
© 2022. The Author(s).
Conflict of interest statement
All authors except N.L.H. were employees of 10x Genomics at the time of submission. Several authors were also shareholders of 10x Genomics at the time of submission. D.B.J., P.S., B.A.A. and W.J.M. are inventors on patent applications assigned to 10x Genomics in relation to algorithms and methods for the study of immune repertoires.
Figures


References
-
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. - PubMed
-
- Heilmann C, Barington T. Distribution of κ and λ light chain isotypes among human blood immunoglobulin-secreting cells after vaccination with pneumococcal polysaccharides. Scand. J. Immunol. 1989;29:159–164. - PubMed
-
- Roy B, et al. High-throughput single-cell analysis of B cell receptor usage among autoantigen-specific plasma cells in celiac disease. J. Immunol. 2017;199:782–791. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

