Emergence of SARS-CoV-2 escape mutations during Bamlanivimab therapy in a phase II randomized clinical trial
- PMID: 36289399
- PMCID: PMC9675946
- DOI: 10.1038/s41564-022-01254-1
Emergence of SARS-CoV-2 escape mutations during Bamlanivimab therapy in a phase II randomized clinical trial
Abstract
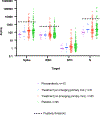
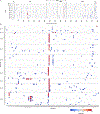
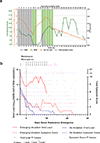
SARS-CoV-2 mutations that cause resistance to monoclonal antibody (mAb) therapy have been reported. However, it remains unclear whether in vivo emergence of SARS-CoV-2 resistance mutations alters viral replication dynamics or therapeutic efficacy in the immune-competent population. As part of the ACTIV-2/A5401 randomized clinical trial (NCT04518410), non-hospitalized participants with symptomatic SARS-CoV-2 infection were given bamlanivimab (700 mg or 7,000 mg) or placebo treatment. Here¸ we report that treatment-emergent resistance mutations [detected through targeted Spike (S) gene next-generation sequencing] were significantly more likely to be detected after bamlanivimab 700 mg treatment compared with the placebo group (7% of 111 vs 0% of 112 participants, P = 0.003). No treatment-emergent resistance mutations among the 48 participants who received 7,000 mg bamlanivimab were recorded. Participants in which emerging mAb resistant virus mutations were identified showed significantly higher pretreatment nasopharyngeal and anterior nasal viral loads. Daily respiratory tract viral sampling through study day 14 showed the dynamic nature of in vivo SARS-CoV-2 infection and indicated a rapid and sustained viral rebound after the emergence of resistance mutations. Participants with emerging bamlanivimab resistance often accumulated additional polymorphisms found in current variants of concern/interest that are associated with immune escape. These results highlight the potential for rapid emergence of resistance during mAb monotherapy treatment that results in prolonged high-level respiratory tract viral loads. Assessment of viral resistance should be prioritized during the development and clinical implementation of antiviral treatments for COVID-19.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing Interests
KWC has received research funding to the institution from Merck Sharpe & Dohme. PK and AN are employees and shareholders of Eli Lilly. ALG reports contract testing from Abbott and research support from Merck and Gilead. ESD has consulted for Gilead, Merck and ViiV and received research support from Gilead and ViiV. ASP has consulted for Amphylx Pharmaceticals. DMS has consulted for Bayer Pharmaceuticals, Linear Therapies, Matrix Biomed, FluxErgy and Brio Clinical. JZL has consulted for Abbvie.
Figures


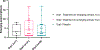



References
-
- Moore CB et al. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296, 1439–1443 (2002). - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
- UM1 AI069423/AI/NIAID NIH HHS/United States
- U54 HL143541/HL/NHLBI NIH HHS/United States
- UM1 AI069424/AI/NIAID NIH HHS/United States
- R01 AI152703/AI/NIAID NIH HHS/United States
- UM1 AI069432/AI/NIAID NIH HHS/United States
- R01 OD011095/OD/NIH HHS/United States
- U01 AI069424/AI/NIAID NIH HHS/United States
- UM1 AI068636/AI/NIAID NIH HHS/United States
- R01 AI028433/AI/NIAID NIH HHS/United States
- UM1 AI068634/AI/NIAID NIH HHS/United States
- UM1 AI106701/AI/NIAID NIH HHS/United States
- R01 AI116868/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

