LAMC2 marks a tumor-initiating cell population with an aggressive signature in pancreatic cancer
- PMID: 36289544
- PMCID: PMC9609288
- DOI: 10.1186/s13046-022-02516-w
LAMC2 marks a tumor-initiating cell population with an aggressive signature in pancreatic cancer
Abstract
Background: Tumor-initiating cells (TIC), also known as cancer stem cells, are considered a specific subpopulation of cells necessary for cancer initiation and metastasis; however, the mechanisms by which they acquire metastatic traits are not well understood.
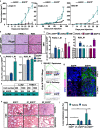
Methods: LAMC2 transcriptional levels were evaluated using publicly available transcriptome data sets, and LAMC2 immunohistochemistry was performed using a tissue microarray composed of PDAC and normal pancreas tissues. Silencing and tracing of LAMC2 was performed using lentiviral shRNA constructs and CRISPR/Cas9-mediated homologous recombination, respectively. The contribution of LAMC2 to PDAC tumorigenicity was explored in vitro by tumor cell invasion, migration, sphere-forming and organoids assays, and in vivo by tumor growth and metastatic assays. mRNA sequencing was performed to identify key cellular pathways upregulated in LAMC2 expressing cells. Metastatic spreading induced by LAMC2- expressing cells was blocked by pharmacological inhibition of transforming growth factor beta (TGF-β) signaling.
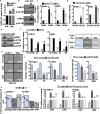
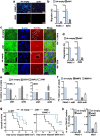
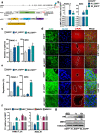
Results: We report a LAMC2-expressing cell population, which is endowed with enhanced self-renewal capacity, and is sufficient for tumor initiation and differentiation, and drives metastasis. mRNA profiling of these cells indicates a prominent squamous signature, and differentially activated pathways critical for tumor growth and metastasis, including deregulation of the TGF-β signaling pathway. Treatment with Vactosertib, a new small molecule inhibitor of the TGF-β type I receptor (activin receptor-like kinase-5, ALK5), completely abrogated lung metastasis, primarily originating from LAMC2-expressing cells.
Conclusions: We have identified a highly metastatic subpopulation of TICs marked by LAMC2. Strategies aimed at targeting the LAMC2 population may be effective in reducing tumor aggressiveness in PDAC patients. Our results prompt further study of this TIC population in pancreatic cancer and exploration as a potential therapeutic target and/or biomarker.
Keywords: Laminin γ2 (LAMC2); Pancreatic ductal adenocarcinoma (PDAC); TGF-β signaling; Tumor-initiating cells (TICs); Vactosertib.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Fitzmaurice C, Global Burden of Disease Cancer Collaboration Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 2006 to 2016: a systematic analysis for the Global Burden of Disease study. J Clin Oncol. 2018;36:1568–1568. - PMC - PubMed
-
- Abel EV, Simeone DM. Biology and clinical applications of pancreatic cancer stem cells. Gastroenterology. 2013;144:1241–1248. - PubMed
-
- Lonardo E, Hermann PC, Mueller M-T, Huber S, Balic A, Miranda-Lorenzo I, et al. Nodal/Activin signaling drives self-renewal and tumorigenicity of pancreatic cancer stem cells and provides a target for combined drug therapy. Cell Stem Cell. 2011;9:433–446. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

