Virtual Screening for SARS-CoV-2 Main Protease Inhibitory Peptides from the Putative Hydrolyzed Peptidome of Rice Bran
- PMID: 36289976
- PMCID: PMC9598432
- DOI: 10.3390/antibiotics11101318
Virtual Screening for SARS-CoV-2 Main Protease Inhibitory Peptides from the Putative Hydrolyzed Peptidome of Rice Bran
Abstract
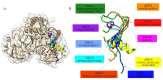
The Coronavirus Disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has led to the loss of life and has affected the life quality, economy, and lifestyle. The SARS-CoV-2 main protease (Mpro), which hydrolyzes the polyprotein, is an interesting antiviral target to inhibit the spreading mechanism of COVID-19. Through predictive digestion, the peptidomes of the four major proteins in rice bran, albumin, glutelin, globulin, and prolamin, with three protease enzymes (pepsin, trypsin, and chymotrypsin), the putative hydrolyzed peptidome was established and used as the input dataset. Then, the prediction of the antiviral peptides (AVPs) was performed by online bioinformatics tools, i.e., AVPpred, Meta-iAVP, AMPfun, and ENNAVIA programs. The amino acid composition and cytotoxicity of candidate AVPs were analyzed by COPid and ToxinPred, respectively. The ten top-ranked antiviral peptides were selected and docked to the SARS-CoV-2 main protease using GalaxyPepDock. Only the top docking scored candidate (AVP4) was further analyzed by molecular dynamics simulation for one nanosecond. According to the bioinformatic analysis results, the candidate SARS-CoV-2 main protease inhibitory peptides were 7-33 amino acid residues and formed hydrogen bonds at Thr22-24, Glu154, and Thr178 in domain 2 with short bonding distances. In addition, these top-ten candidate bioactive peptides contain hydrophilic amino acid residues and have a positive net charge. We hope that this study will provide a potential starting point for peptide-based therapeutic agents against COVID-19.
Keywords: SARS-CoV-2 main protease; antiviral peptide; bioinformatics; molecular docking; molecular dynamics; rice bran.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Computer-Aided Screening for Potential Coronavirus 3-Chymotrypsin-like Protease (3CLpro) Inhibitory Peptides from Putative Hemp Seed Trypsinized Peptidome.Molecules. 2022 Dec 21;28(1):50. doi: 10.3390/molecules28010050. Molecules. 2022. PMID: 36615263 Free PMC article.
-
Identification of natural antimicrobial peptides mimetic to inhibit Ca2+ influx DDX3X activity for blocking dengue viral infectivity.J Bioenerg Biomembr. 2024 Apr;56(2):125-139. doi: 10.1007/s10863-023-09996-1. Epub 2023 Dec 14. J Bioenerg Biomembr. 2024. PMID: 38095733
-
In silico validation of coumarin derivatives as potential inhibitors against Main Protease, NSP10/NSP16-Methyltransferase, Phosphatase and Endoribonuclease of SARS CoV-2.J Biomol Struct Dyn. 2021 Nov;39(18):7306-7321. doi: 10.1080/07391102.2020.1808075. Epub 2020 Aug 24. J Biomol Struct Dyn. 2021. PMID: 32835632 Free PMC article.
-
Identification of new anti-nCoV drug chemical compounds from Indian spices exploiting SARS-CoV-2 main protease as target.J Biomol Struct Dyn. 2021 Jun;39(9):3428-3434. doi: 10.1080/07391102.2020.1763202. Epub 2020 May 13. J Biomol Struct Dyn. 2021. PMID: 32362243 Free PMC article.
-
A Potential Peptide From Soy Cheese Produced Using Lactobacillus delbrueckii WS4 for Effective Inhibition of SARS-CoV-2 Main Protease and S1 Glycoprotein.Front Mol Biosci. 2020 Dec 11;7:601753. doi: 10.3389/fmolb.2020.601753. eCollection 2020. Front Mol Biosci. 2020. PMID: 33363209 Free PMC article.
Cited by
-
Marine Brown Algae-Derived Compounds as Potential Inhibitors of Japanese Encephalitis Virus RNA-Dependent RNA Polymerase.Mar Drugs. 2024 Feb 17;22(2):92. doi: 10.3390/md22020092. Mar Drugs. 2024. PMID: 38393063 Free PMC article.
-
Virtual Screening of Peptide Libraries: The Search for Peptide-Based Therapeutics Using Computational Tools.Int J Mol Sci. 2024 Feb 1;25(3):1798. doi: 10.3390/ijms25031798. Int J Mol Sci. 2024. PMID: 38339078 Free PMC article. Review.
-
Olive Leaf Protein Hydrolysate as a Novel Source of Antimicrobial Peptides: Peptidomic Characterization and In Silico Evaluation.Molecules. 2025 Aug 14;30(16):3382. doi: 10.3390/molecules30163382. Molecules. 2025. PMID: 40871537 Free PMC article.
-
Computer-Aided Screening for Potential Coronavirus 3-Chymotrypsin-like Protease (3CLpro) Inhibitory Peptides from Putative Hemp Seed Trypsinized Peptidome.Molecules. 2022 Dec 21;28(1):50. doi: 10.3390/molecules28010050. Molecules. 2022. PMID: 36615263 Free PMC article.
-
Computer-Aided Virtual Screening and In Vitro Validation of Biomimetic Tyrosinase Inhibitory Peptides from Abalone Peptidome.Int J Mol Sci. 2023 Feb 5;24(4):3154. doi: 10.3390/ijms24043154. Int J Mol Sci. 2023. PMID: 36834568 Free PMC article.
References
-
- Li X., Okita T.W. Accumulation of Prolamines and Glutelins during Rice Seed Development: A Quantitative Evaluation. Plant Cell Physiol. 1993;34:385–390.
LinkOut - more resources
Full Text Sources
Miscellaneous

