Contribution of Symptomatic, Herbal Treatment Options to Antibiotic Stewardship and Microbiotic Health
- PMID: 36289988
- PMCID: PMC9598931
- DOI: 10.3390/antibiotics11101331
Contribution of Symptomatic, Herbal Treatment Options to Antibiotic Stewardship and Microbiotic Health
Abstract
Epithelial surfaces in humans are home to symbiotic microbes (i.e., microbiota) that influence the defensive function against pathogens, depending on the health of the microbiota. Healthy microbiota contribute to the well-being of their host, in general (e.g., via the gut-brain axis), and their respective anatomical site, in particular (e.g., oral, urogenital, skin, or respiratory microbiota). Despite efforts towards a more responsible use of antibiotics, they are often prescribed for uncomplicated, self-limiting infections and can have a substantial negative impact on the gut microbiota. Treatment alternatives, such as non-steroidal anti-inflammatory drugs, may also influence the microbiota; thus, they can have lasting adverse effects. Herbal drugs offer a generally safe treatment option for uncomplicated infections of the urinary or respiratory tract. Additionally, their microbiota preserving properties allow for a more appropriate therapy of uncomplicated infections, without contributing to an increase in antibiotic resistance or disturbing the gut microbiota. Here, herbal treatments may be a more appropriate therapy, with a generally favorable safety profile.
Keywords: NSAID; antibiotic stewardship; gut microbiota; herbal drugs; homeostasis; uncomplicated infection.
Conflict of interest statement
B Nausch, CB Bittner, M Höller, and D Abramov-Sommariva are employees of Bionorica SE. A. Gessner receives consultancy fees from Bionorica SE.
Figures
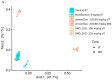

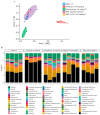
References
-
- Vaga S., Lee S., Ji B., Andreasson A., Talley N.J., Agréus L., Bidkhori G., Kovatcheva-Datchary P., Park J., Lee D., et al. Compositional and functional differences of the mucosal microbiota along the intestine of healthy individuals. Sci. Rep. 2020;10:14977. doi: 10.1038/s41598-020-71939-2. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

