Machine-Learning-Assisted Analysis of TCR Profiling Data Unveils Cross-Reactivity between SARS-CoV-2 and a Wide Spectrum of Pathogens and Other Diseases
- PMID: 36290433
- PMCID: PMC9598299
- DOI: 10.3390/biology11101531
Machine-Learning-Assisted Analysis of TCR Profiling Data Unveils Cross-Reactivity between SARS-CoV-2 and a Wide Spectrum of Pathogens and Other Diseases
Abstract
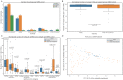
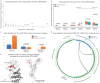
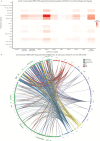
During the last two years, the emergence of SARS-CoV-2 has led to millions of deaths worldwide, with a devastating socio-economic impact on a global scale. The scientific community's focus has recently shifted towards the association of the T cell immunological repertoire with COVID-19 progression and severity, by utilising T cell receptor sequencing (TCR-Seq) assays. The Multiplexed Identification of T cell Receptor Antigen (MIRA) dataset, which is a subset of the immunoACCESS study, provides thousands of TCRs that can specifically recognise SARS-CoV-2 epitopes. Our study proposes a novel Machine Learning (ML)-assisted approach for analysing TCR-Seq data from the antigens' point of view, with the ability to unveil key antigens that can accurately distinguish between MIRA COVID-19-convalescent and healthy individuals based on differences in the triggered immune response. Some SARS-CoV-2 antigens were found to exhibit equal levels of recognition by MIRA TCRs in both convalescent and healthy cohorts, leading to the assumption of putative cross-reactivity between SARS-CoV-2 and other infectious agents. This hypothesis was tested by combining MIRA with other public TCR profiling repositories that host assays and sequencing data concerning a plethora of pathogens. Our study provides evidence regarding putative cross-reactivity between SARS-CoV-2 and a wide spectrum of pathogens and diseases, with M. tuberculosis and Influenza virus exhibiting the highest levels of cross-reactivity. These results can potentially shift the emphasis of immunological studies towards an increased application of TCR profiling assays that have the potential to uncover key mechanisms of cell-mediated immune response against pathogens and diseases.
Keywords: COVID-19; MIRA dataset; Machine Learning; T cell receptor; cross-reactivity phenomenon; diseases; pathogens.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- JHCRC . John Hopkins Coronavirus Resource Center. Johns Hopkins University School of Medicine; Baltimore, MD, USA: [(accessed on 23 September 2022)]. Available online: https://coronavirus.jhu.edu/covid-19-daily-video.
-
- Eastin C., Eastin T. Clinical Characteristics of Coronavirus Disease 2019 in China. J. Emerg. Med. 2020;58:711–712. doi: 10.1016/j.jemermed.2020.04.004. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

