Identification of the Factor That Leads Human Mesenchymal Stem Cell Lines into Decellularized Bone
- PMID: 36290460
- PMCID: PMC9598111
- DOI: 10.3390/bioengineering9100490
Identification of the Factor That Leads Human Mesenchymal Stem Cell Lines into Decellularized Bone
Abstract
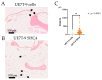
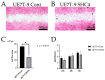
Hematopoiesis is maintained by the interaction of hematopoietic stem cells (HSCs) and bone marrow mesenchymal stem cells (MSCs) in bone marrow microenvironments, called niches. Certain genetic mutations in MSCs, not HSCs, provoke some hematopoietic neoplasms, such as myelodysplastic syndrome. An in vivo bone marrow niche model using human MSC cell lines with specific genetic mutations and bone scaffolds is necessary to elucidate these interactions and the disease onset. We focused on decellularized bone (DCB) as a useful bone scaffold and attempted to induce human MSCs (UE7T-9 cells) into the DCB. Using the CRISPR activation library, we identified SHC4 upregulation as a candidate factor, with the SHC4 overexpression in UE7T-9 cells activating their migratory ability and upregulating genes to promote hematopoietic cell migration. This is the first study to apply the CRISPR library to engraft cells into decellularized biomaterials. SHC4 overexpression is essential for engrafting UE7T-9 cells into DCB, and it might be the first step toward creating an in vivo human-mouse hybrid bone marrow niche model.
Keywords: CRISPR SAM activation library; bone marrow niche; decellularized bone marrow; mesenchymal stem cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Reconstitution of bone-like matrix in osteogenically differentiated mesenchymal stem cell-collagen constructs: A three-dimensional in vitro model to study hematopoietic stem cell niche.J Tissue Eng. 2013 Oct 10;4:2041731413508668. doi: 10.1177/2041731413508668. eCollection 2013. J Tissue Eng. 2013. PMID: 24555007 Free PMC article.
-
Biomimetic macroporous PEG hydrogels as 3D scaffolds for the multiplication of human hematopoietic stem and progenitor cells.Biomaterials. 2014 Jan;35(3):929-40. doi: 10.1016/j.biomaterials.2013.10.038. Epub 2013 Oct 28. Biomaterials. 2014. PMID: 24176196
-
Bone Marrow Niches of Hematopoietic Stem and Progenitor Cells.Int J Mol Sci. 2022 Apr 18;23(8):4462. doi: 10.3390/ijms23084462. Int J Mol Sci. 2022. PMID: 35457280 Free PMC article. Review.
-
The bone marrow niche: habitat to hematopoietic and mesenchymal stem cells, and unwitting host to molecular parasites.Leukemia. 2008 May;22(5):941-50. doi: 10.1038/leu.2008.48. Epub 2008 Feb 28. Leukemia. 2008. PMID: 18305549 Free PMC article. Review.
-
[Bone and marrow niches for hematopoiesis].Clin Calcium. 2012 Nov;22(11):1659-67. Clin Calcium. 2012. PMID: 23103809 Review. Japanese.
Cited by
-
Unlocking the Potential of Stem Cell Microenvironments In Vitro.Bioengineering (Basel). 2024 Mar 19;11(3):289. doi: 10.3390/bioengineering11030289. Bioengineering (Basel). 2024. PMID: 38534563 Free PMC article. Review.
References
-
- Baccin C., Al-Sabah J., Velten L., Helbling P.M., Grünschläger F., Hernández-Malmierca P., Nombela-Arrieta C., Steinmetz L.M., Trumpp A., Haas S. Combined single-cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nat. Cell Biol. 2020;22:38–48. doi: 10.1038/s41556-019-0439-6. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

