Transcriptomic Analysis of Genes Associated with Oxidative Stress in Chronic Rhinosinusitis Patients with Nasal Polyps: Identifying Novel Genes Involved in Nasal Polyposis
- PMID: 36290622
- PMCID: PMC9598890
- DOI: 10.3390/antiox11101899
Transcriptomic Analysis of Genes Associated with Oxidative Stress in Chronic Rhinosinusitis Patients with Nasal Polyps: Identifying Novel Genes Involved in Nasal Polyposis
Abstract
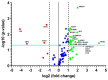
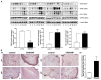
Chronic rhinosinusitis with nasal polyps (CRSwNP) is a complicated inflammatory disease, and the underlying mechanism remains unclear. While some reactive oxygen/nitrogen species-related gene products are reported to participate in CRSwNP, a systemic and full analysis of oxidative-stress-associated genes in CRSwNP has not been extensively studied. Therefore, this study sought to catalog the gene-expression patterns related to oxidative stress and antioxidant defense in control and CRSwNP patients. In total, 25 control and 25 CRSwNP patients were recruited. The distribution and expression of 4-hydroxynonenal and 3-nitrotyrosine as markers of oxidative stress-which is represented by lipid peroxidation and the protein nitration of tyrosine residues in CRSwNP nasal polyps (NPs)-were more apparently increased than those found in the control nasal mucosae, as determined by immunohistochemistry (IHC). The expression of 84 oxidative-stress-related genes in nasal mucosae and NP tissues was analyzed via real-time PCR, which showed that 19 genes and 4 genes were significantly up- and downregulated, respectively; among them, inducible nitric oxide synthase (iNOS) and heme oxygenase 1 (HO-1) were notably upregulated, whereas lactoperoxidase (LPO), myeloperoxidase (MPO), and superoxide dismutase 3 (SOD3) were highly downregulated. Changes in the mRNA and protein levels of these redox proteins were confirmed with a customized, real-time PCR array and RT-PCR analysis, as well as Western blotting and IHC assays. A receiver operating characteristic curve analysis further suggested that LPO, MPO, SOD3, HO-1, and iNOS are possible endotype predictors of CRSwNP development. Collectively, we present an oxidative-stress-related gene profile of CRSwNP NP tissues, providing evidence that the systemic changes in oxidative stress and the antioxidative defense system, including novel iNOS, heme peroxidases, and other genes, are closely linked to CRSwNP pathology, development, and progression.
Keywords: CRS; CRSwNP; ROS; lactoperoxidase; oxidative stress; transcriptional profiling.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analysis, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
Figures




Similar articles
-
Increased Expression of TXNIP Facilitates Oxidative Stress in Nasal Epithelial Cells of Patients With Chronic Rhinosinusitis With Nasal Polyps.Am J Rhinol Allergy. 2021 Sep;35(5):607-614. doi: 10.1177/1945892420982411. Epub 2020 Dec 29. Am J Rhinol Allergy. 2021. PMID: 33375816
-
Genetic polymorphism of antioxidant enzymes in eosinophilic and non-eosinophilic nasal polyposis.Eur Arch Otorhinolaryngol. 2017 Jan;274(1):267-273. doi: 10.1007/s00405-016-4259-z. Epub 2016 Aug 11. Eur Arch Otorhinolaryngol. 2017. PMID: 27515707
-
Expression of heme oxygenase-1 in eosinophilic and non-eosinophilic chronic rhinosinusitis with nasal polyps: modulation by cytokines.Int Forum Allergy Rhinol. 2015 Aug;5(8):734-40. doi: 10.1002/alr.21530. Epub 2015 Apr 23. Int Forum Allergy Rhinol. 2015. PMID: 25907676
-
Systematic literature review of humanistic and economic burdens of chronic rhinosinusitis with nasal polyposis.Curr Med Res Opin. 2020 Nov;36(11):1913-1926. doi: 10.1080/03007995.2020.1815683. Epub 2020 Sep 25. Curr Med Res Opin. 2020. PMID: 32851882
-
The Effect of the Premedication with Systemic Corticosteroids and Antibiotics on Inflammation and Intraoperative Bleeding During Sinonasal Endoscopic Surgery for Chronic Rhinosinusitis with Nasal Polyps (CRSwNP).J Craniofac Surg. 2022 Jun 1;33(4):e405-e408. doi: 10.1097/SCS.0000000000008285. Epub 2022 Jan 28. J Craniofac Surg. 2022. PMID: 35093993
Cited by
-
Serum exosomal miR-141-3p and miR-3679-5p levels associated with endotype and postoperative recurrence in chronic rhinosinusitis with nasal polyps.World Allergy Organ J. 2024 Jul 24;17(8):100938. doi: 10.1016/j.waojou.2024.100938. eCollection 2024 Aug. World Allergy Organ J. 2024. PMID: 39156601 Free PMC article.
-
Oxidative stress and reactive oxygen species in otorhinolaryngological diseases: insights from pathophysiology to targeted antioxidant therapies.Redox Rep. 2025 Dec;30(1):2458942. doi: 10.1080/13510002.2025.2458942. Epub 2025 Feb 2. Redox Rep. 2025. PMID: 39894944 Free PMC article. Review.
-
Supersulfide formation in the sinus mucosa of chronic rhinosinusitis.Laryngoscope Investig Otolaryngol. 2024 Jul 27;9(4):e1261. doi: 10.1002/lio2.1261. eCollection 2024 Aug. Laryngoscope Investig Otolaryngol. 2024. PMID: 39071205 Free PMC article.
-
Effect of cigarette smoke on the proliferation, viability, gene expression, and cellular functions of adipose-derived mesenchymal stem cells from smoking and non-smoking donors.Biol Open. 2024 Dec 15;13(12):bio061665. doi: 10.1242/bio.061665. Epub 2024 Dec 3. Biol Open. 2024. PMID: 39625294 Free PMC article.
-
SLC27A2 marks lipid peroxidation in nasal epithelial cells driven by type 2 inflammation in chronic rhinosinusitis with nasal polyps.Exp Mol Med. 2025 Apr;57(4):856-871. doi: 10.1038/s12276-025-01440-1. Epub 2025 Apr 7. Exp Mol Med. 2025. PMID: 40195539 Free PMC article.
References
-
- Wang X., Zhang N., Bo M., Holtappels G., Zheng M., Lou H., Wang H., Zhang L., Bachert C. Diversity of T(H) cytokine profiles in patients with chronic rhinosinusitis: A multicenter study in Europe, Asia, and Oceania. J. Allergy Clin. Immunol. 2016;138:1344–1353. doi: 10.1016/j.jaci.2016.05.041. - DOI - PubMed
-
- Takabayashi T., Kato A., Peters A.T., Hulse K.E., Suh L.A., Carter R., Norton J., Grammer L.C., Cho S.H., Tan B.K., et al. Excessive fibrin deposition in nasal polyps caused by fibrinolytic impairment through reduction of tissue plasminogen activator expression. Am. J. Respir. Crit. Care Med. 2013;187:49–57. doi: 10.1164/rccm.201207-1292OC. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

