Super-Long SERS Active Single Silver Nanowires for Molecular Imaging in 2D and 3D Cell Culture Models
- PMID: 36291012
- PMCID: PMC9599576
- DOI: 10.3390/bios12100875
Super-Long SERS Active Single Silver Nanowires for Molecular Imaging in 2D and 3D Cell Culture Models
Abstract
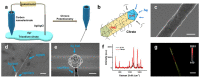
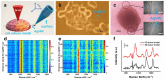
Establishing a systematic molecular information analysis strategy for cell culture models is of great significance for drug development and tissue engineering technologies. Here, we fabricated single silver nanowires with high surface-enhanced Raman scattering activity to extract SERS spectra in situ from two-dimensional (2D) and three-dimensional (3D) cell culture models. The silver nanowires were super long, flexible and thin enough to penetrate through multiple cells. A single silver nanowire was used in combination with a four-dimensional microcontroller as a cell endoscope for spectrally analyzing the components in cell culture models. Then, we adopted a machine learning algorithm to analyze the obtained spectra. Our results show that the abundance of proteins differs significantly between the 2D and 3D models, and that nucleic acid-rich and protein-rich regions can be distinguished with satisfactory accuracy.
Keywords: Ag nanowire; SERS; cell culture model; machine learning; spatial resolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

