Exosomal Plasma Gelsolin Is an Immunosuppressive Mediator in the Ovarian Tumor Microenvironment and a Determinant of Chemoresistance
- PMID: 36291171
- PMCID: PMC9600545
- DOI: 10.3390/cells11203305
Exosomal Plasma Gelsolin Is an Immunosuppressive Mediator in the Ovarian Tumor Microenvironment and a Determinant of Chemoresistance
Abstract
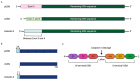
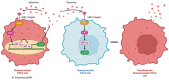
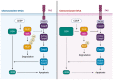
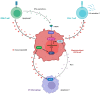
Ovarian Cancer (OVCA) is the most fatal gynecologic cancer and has a 5-year survival rate less than 45%. This is mainly due to late diagnosis and drug resistance. Overexpression of plasma gelsolin (pGSN) is key contributing factor to OVCA chemoresistance and immunosuppression. Gelsolin (GSN) is a multifunctional protein that regulates the activity of actin filaments by cleavage, capping, and nucleation. Generally, it plays an important role in cytoskeletal remodeling. GSN has three isoforms: cytosolic GSN, plasma GSN (pGSN), and gelsolin-3. Exosomes containing pGSN are released and contribute to the progression of OVCA. This review describes how pGSN overexpression inhibits chemotherapy-induced apoptosis and triggers positive feedback loops of pGSN expression. It also describes the mechanisms by which exosomal pGSN promotes apoptosis and dysfunction in tumor-killing immune cells. A discussion on the potential of pGSN as a prognostic, diagnostic, and therapeutic marker is also presented herein.
Keywords: T cells; apoptosis; chemoresistance; extracellular vesicles (EVs); immune cells; macrophages; ovarian cancer; plasma gelsolin (pGSN); tumor microenvironment (TME).
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Surveillance, Epidemiology, and End Results (SEER) Program. SEER*Stat Database: Incidence—SEER Research Data, 8 Registries, Nov 2021 Sub (1975–2019)—Linked to County Attributes—Time Dependent (1990–2019) Income/Rurality, 1969–2020 Counties, National Cancer Institute, DCCPS, Surveillance Research Program, Released April 2022, Based on the November 2021 Submission. [(accessed on 26 August 2022)]; Available online: https://seer.cancer.gov/statfacts/html/cervix.html.
-
- Surveillance, Epidemiology, and End Results (SEER) Program. SEER*Stat Database: Incidence—SEER Research Data, 8 Registries, Nov 2021 Sub (1975–2019)—Linked to County Attributes—Time Dependent (1990–2019) Income/Rurality, 1969–2020 Counties, National Cancer Institute, DCCPS, Surveillance Research Program, Released April 2022, Based on the November 2021 Submission. [(accessed on 26 August 2022)]; Available online: https://seer.cancer.gov/statfacts/html/corp.html.
-
- Surveillance, Epidemiology, and End Results (SEER) Program. SEER*Stat Database: Incidence—SEER Research Data, 8 Registries, Nov 2021 Sub (1975–2019)—Linked to County Attributes—Time Dependent (1990–2019) Income/Rurality, 1969–2020 Counties, National Cancer Institute, DCCPS, Surveillance Research Program, Released April 2022, Based on the November 2021 Submission. [(accessed on 26 August 2022)]; Available online: https://seer.cancer.gov/statfacts/html/ovary.html.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

