Dissecting the Immunological Profiles in NSD3-Amplified LUSC through Integrative Multi-Scale Analyses
- PMID: 36291782
- PMCID: PMC9599511
- DOI: 10.3390/cancers14204997
Dissecting the Immunological Profiles in NSD3-Amplified LUSC through Integrative Multi-Scale Analyses
Abstract
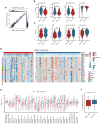
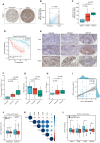
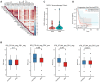
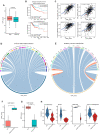
The histone H3 lysine 36 (H3K36) methyltransferase NSD3, a neighboring gene of FGFR1, has been identified as a critical genetic driver of lung squamous cell carcinoma (LUSC). However, the molecular characteristics, especially the immunological roles of NSD3 in driving carcinogenesis, are poorly understood. In this study, we systematically integrated multi-omics data (e.g., genome, transcriptome, proteome, and TMA array) to dissect the immunological profiles in NSD3-amplified LUSC. Next, pharmaco-transcriptomic correlation analysis was implemented to identify the molecular underpinnings and therapeutic vulnerabilities in LUSC. We revealed that NSD3-amplified LUSC presents a non-inflamed tumor immune microenvironment (TIME) state in multiple independent LUSC patient cohorts. Predictably, elevated NSD3 expression was correlated with a worse immunotherapy outcome. Further molecular characterizations revealed that the high activity of unfolded protein response (UPR) signaling might be a pivotal mediator for the non-immunogenic phenotype of NSD3-amplified LUSC. Concordantly, we showed that NSD3-amplified LUSCs exhibited a more sensitive phenotype to compounds targeting UPR branches than the wild-type group. In brief, our multi-level analyses point to a previously unappreciated immunological role for NSD3 and provide therapeutic rationales for NSD3-amplified squamous lung cancer.
Keywords: NSD3; immunotherapy; lung squamous cell carcinoma (LUSC); tumor immune microenvironment (TIME); unfolded protein response (UPR).
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Kim Y., Hammerman P.S., Kim J., Yoon J.-A., Lee Y., Sun J.-M., Wilkerson M.D., Pedamallu C.S., Cibulskis K., Yoo Y.K., et al. Integrative and Comparative Genomic Analysis of Lung Squamous Cell Carcinomas in East Asian Patients. J. Clin. Oncol. 2014;32:121–128. doi: 10.1200/JCO.2013.50.8556. - DOI - PMC - PubMed
-
- Campbell J.D., Alexandrov A., Kim J., Wala J., Berger A.H., Pedamallu C.S., Shukla S.A., Guo G., Brooks A.N., Murray B.A., et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat. Genet. 2016;48:607–616. doi: 10.1038/ng.3564. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

