CSMD1 Shows Complex Patterns of Somatic Copy Number Alterations and Expressions of mRNAs and Target Micro RNAs in Esophageal Squamous Cell Carcinoma
- PMID: 36291785
- PMCID: PMC9599939
- DOI: 10.3390/cancers14205001
CSMD1 Shows Complex Patterns of Somatic Copy Number Alterations and Expressions of mRNAs and Target Micro RNAs in Esophageal Squamous Cell Carcinoma
Abstract
Background: Human Cub and Sushi Multiple Domains 1 (CSMD1) is a novel candidate tumor-suppressor gene that codes for multiple domains, including complement regulatory and adhesion proteins, and has recently been shown to have alterations in multiple cancers. We investigated CSMD1 in esophageal squamous cell carcinoma (ESCC) by performing an integrated analysis on somatic copy number alterations (CNAs), including copy-number gain or loss, allelic imbalance (AI), loss of heterozygosity (LOH), and the expressions of mRNA and its target miRNAs on specimens from the same patients with ESCC.
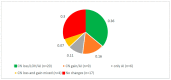
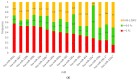
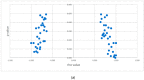
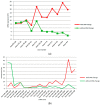
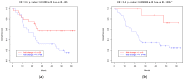
Results: (i) Two-thirds of ESCC patients had all three types of alterations studied-somatic DNA alterations in 70%, and abnormal expressions of CSMD1 RNA in 69% and in target miRNAs in 66%; patterns among these alterations were complex. (ii) In total, 97% of 888 CSMD1 SNPs studied showed somatic DNA alterations, with most located near exons 4-11, 24-25, 39-40, 55-56, and 69-70. (iii) In total, 68% of SNPs with a CNA were correlated with expression of CSMD1. (iv) A total of 33 correlations between non-coding SNPs and expression of CSMD1 target miRs were found.
Conclusions: Our results indicate that the CSMD1 gene may play a role in ESCC through complex patterns of DNA alterations and RNA and miRNA expressions. Alterations in some somatic SNPs in non-coding regions of CSMD1 appear to influence expression of this gene and its target miRNAs.
Keywords: CSMD1; allelic imbalance; esophageal squamous cell carcinoma; gene expression; miRNA; somatic copy number alternation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Kamal M., Shaaban A.M., Zhang L., Walker C., Gray S., Thakker N., Toomes C., Speirs V., Bell S.M. Loss of CSMD1 expression is associated with high tumour grade and poor survival in invasive ductal breast carcinoma. Breast Cancer Res. Treat. 2010;121:555–563. doi: 10.1007/s10549-009-0500-4. - DOI - PubMed
-
- Escudero-Esparza A., Kalchishkova N., Kurbasic E., Jiang W.G., Blom A.M. The novel complement inhibitor human CUB and Sushi multiple domains 1 (CSMD1) protein promotes factor I-mediated degradation of C4b and C3b and inhibits the membrane attack complex assembly. FASEB J. 2013;27:5083–5093. doi: 10.1096/fj.13-230706. - DOI - PubMed
-
- Kraus D.M., Elliott G.S., Chute H., Horan T., Pfenninger K.H., Sanford S.D., Foster S., Scully S., Welcher A.A., Holers V.M. CSMD1 is a novel multiple domain complement-regulatory protein highly expressed in the central nervous system and epithelia tissues. J. Immunol. 2006;176:4419–4430. doi: 10.4049/jimmunol.176.7.4419. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

