Application of CRISPR/Cas Systems in the Nucleic Acid Detection of Infectious Diseases
- PMID: 36292145
- PMCID: PMC9600689
- DOI: 10.3390/diagnostics12102455
Application of CRISPR/Cas Systems in the Nucleic Acid Detection of Infectious Diseases
Abstract
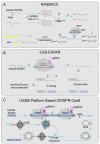
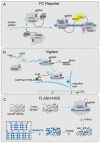
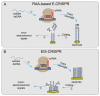
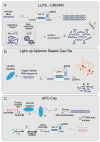
The CRISPR/Cas system is a protective adaptive immune system against attacks from foreign mobile genetic elements. Since the discovery of the excellent target-specific sequence recognition ability of the CRISPR/Cas system, the CRISPR/Cas system has shown excellent performance in the development of pathogen nucleic-acid-detection technology. In combination with various biosensing technologies, researchers have made many rapid, convenient, and feasible innovations in pathogen nucleic-acid-detection technology. With an in-depth understanding and development of the CRISPR/Cas system, it is no longer limited to CRISPR/Cas9, CRISPR/Cas12, and other systems that had been widely used in the past; other CRISPR/Cas families are designed for nucleic acid detection. We summarized the application of CRISPR/Cas-related technology in infectious-disease detection and its development in SARS-CoV-2 detection.
Keywords: CRISPR/Cas system; SARS-CoV-2; biosensing technologies; diagnoses; pathogen nucleic acids.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
CRISPR-cas technology: A key approach for SARS-CoV-2 detection.Front Bioeng Biotechnol. 2023 Apr 12;11:1158672. doi: 10.3389/fbioe.2023.1158672. eCollection 2023. Front Bioeng Biotechnol. 2023. PMID: 37214290 Free PMC article. Review.
-
Advances in the application of CRISPR-Cas technology in rapid detection of pathogen nucleic acid.Front Mol Biosci. 2023 Sep 21;10:1260883. doi: 10.3389/fmolb.2023.1260883. eCollection 2023. Front Mol Biosci. 2023. PMID: 37808520 Free PMC article. Review.
-
Development of CRISPR-Mediated Nucleic Acid Detection Technologies and Their Applications in the Livestock Industry.Genes (Basel). 2022 Nov 2;13(11):2007. doi: 10.3390/genes13112007. Genes (Basel). 2022. PMID: 36360244 Free PMC article. Review.
-
CRISPR technology incorporating amplification strategies: molecular assays for nucleic acids, proteins, and small molecules.Chem Sci. 2021 Mar 2;12(13):4683-4698. doi: 10.1039/d0sc06973f. Chem Sci. 2021. PMID: 34163728 Free PMC article. Review.
-
CRISPR-Cas-based techniques for pathogen detection: Retrospect, recent advances, and future perspectives.J Adv Res. 2023 Aug;50:69-82. doi: 10.1016/j.jare.2022.10.011. Epub 2022 Oct 30. J Adv Res. 2023. PMID: 36367481 Free PMC article. Review.
Cited by
-
Advancements and prospects of CRISPR/Cas9 technologies for abiotic and biotic stresses in sugar beet.Front Genet. 2023 Nov 9;14:1235855. doi: 10.3389/fgene.2023.1235855. eCollection 2023. Front Genet. 2023. PMID: 38028586 Free PMC article. Review.
-
Amplification-Based CRISPR/Cas12a Biosensor Targeting the COX1 Gene for Specific Detection of Porcine DNA.ACS Omega. 2023 Oct 8;8(41):38212-38219. doi: 10.1021/acsomega.3c04473. eCollection 2023 Oct 17. ACS Omega. 2023. PMID: 37867655 Free PMC article.
-
Cellular resistance to an oncolytic virus is driven by chronic activation of innate immunity.iScience. 2022 Dec 7;26(1):105749. doi: 10.1016/j.isci.2022.105749. eCollection 2023 Jan 20. iScience. 2022. PMID: 36590165 Free PMC article.
-
Engineered Electrochemiluminescence Biosensors for Monitoring Heavy Metal Ions: Current Status and Prospects.Biosensors (Basel). 2023 Dec 22;14(1):9. doi: 10.3390/bios14010009. Biosensors (Basel). 2023. PMID: 38248386 Free PMC article. Review.
-
The application of CRISPR-Cas system in Staphylococcus aureus infection.Heliyon. 2024 Jul 10;10(14):e34383. doi: 10.1016/j.heliyon.2024.e34383. eCollection 2024 Jul 30. Heliyon. 2024. PMID: 39108851 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

