Characterization of the Chloroplast Genome of Argyranthemum frutescens and a Comparison with Other Species in Anthemideae
- PMID: 36292605
- PMCID: PMC9602088
- DOI: 10.3390/genes13101720
Characterization of the Chloroplast Genome of Argyranthemum frutescens and a Comparison with Other Species in Anthemideae
Abstract
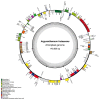
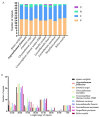
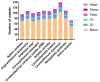
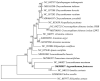
Argyranthemum frutescens, which belongs to the Anthemideae (Asteraceae), is widely cultivated as an ornamental plant. In this study, the complete chloroplast genome of A. frutescens was obtained based on the sequences generated by Illumina HiSeq. The chloroplast genome of A. frutescens was 149,626 base pairs (bp) in length, containing a pair of inverted repeats (IR, 24,510 bp) regions separated by a small single-copy (SSC, 18,352 bp) sequence and a large single-copy (LSC, 82,254 bp) sequence. The genome contained 132 genes, consisting of 85 coding DNA sequences, 37 tRNA genes, and 8 rRNA genes, with nineteen genes duplicated in the IR region. A comparison chloroplast genome analysis among ten species from the tribe of Anthemideae revealed that the chloroplast genome size varied, but the genome structure, gene content, and oligonucleotide repeats were highly conserved. Highly divergent regions, e.g., ycf1, trnK-psbK, petN-psbM intronic, were detected. Phylogenetic analysis supported Argyranthemum as a separate genus. The findings of this study will be helpful in the exploration of the phylogenetic relationships of the tribe of Anthemideae and contribute to the breeding improvement of A. frutescens.
Keywords: Anthemideae; Argyranthemum frutescens; chloroplast genome; ornamental plant; phylogenetic relationship.
Conflict of interest statement
There are no conflict of interest to declare.
Figures

Similar articles
-
Complete Chloroplast Genome Sequence of the Long Blooming Cultivar Camellia 'Xiari Qixin': Genome Features, Comparative and Phylogenetic Analysis.Genes (Basel). 2023 Feb 10;14(2):460. doi: 10.3390/genes14020460. Genes (Basel). 2023. PMID: 36833387 Free PMC article.
-
Complete chloroplast genome sequences of Mongolia medicine Artemisia frigida and phylogenetic relationships with other plants.PLoS One. 2013;8(2):e57533. doi: 10.1371/journal.pone.0057533. Epub 2013 Feb 27. PLoS One. 2013. PMID: 23460871 Free PMC article.
-
The complete chloroplast genome sequence of Perilla frutescens (L.).Mitochondrial DNA A DNA Mapp Seq Anal. 2016 Sep;27(5):3306-7. doi: 10.3109/19401736.2015.1015015. Epub 2015 Feb 25. Mitochondrial DNA A DNA Mapp Seq Anal. 2016. PMID: 25714143
-
Comparative analysis of complete chloroplast genomes of 14 Asteraceae species.Mol Biol Rep. 2024 Oct 26;51(1):1094. doi: 10.1007/s11033-024-10030-9. Mol Biol Rep. 2024. PMID: 39460814
-
Comparative genomics of four Liliales families inferred from the complete chloroplast genome sequence of Veratrum patulum O. Loes. (Melanthiaceae).Gene. 2013 Nov 10;530(2):229-35. doi: 10.1016/j.gene.2013.07.100. Epub 2013 Aug 23. Gene. 2013. PMID: 23973725
Cited by
-
Chloroplast genome of four Amorphophallus species: genomic features,comparative analysis, and phylogenetic relationships among Amorphophallus species.BMC Genomics. 2024 Nov 21;25(1):1122. doi: 10.1186/s12864-024-11053-z. BMC Genomics. 2024. PMID: 39567899 Free PMC article.
-
Complete Chloroplast Genome Sequence of the Long Blooming Cultivar Camellia 'Xiari Qixin': Genome Features, Comparative and Phylogenetic Analysis.Genes (Basel). 2023 Feb 10;14(2):460. doi: 10.3390/genes14020460. Genes (Basel). 2023. PMID: 36833387 Free PMC article.
-
The chloroplast genome sequence and phylogenetic analysis of Rubia alata Wall and Rubia ovatifolia Z. Ying Zhang. (Rubiaceae).Mol Biol Rep. 2024 Nov 11;51(1):1140. doi: 10.1007/s11033-024-10046-1. Mol Biol Rep. 2024. PMID: 39527330
-
Complete sequence of the Achillea ptarmica chloroplast genome determined by long-read sequencing.Mitochondrial DNA B Resour. 2025 Jul 23;10(8):753-757. doi: 10.1080/23802359.2025.2535628. eCollection 2025. Mitochondrial DNA B Resour. 2025. PMID: 40708795 Free PMC article.
-
Complete chloroplast genome sequence of Camellia sinensis: genome structure, adaptive evolution, and phylogenetic relationships.J Appl Genet. 2023 Sep;64(3):419-429. doi: 10.1007/s13353-023-00767-7. Epub 2023 Jun 28. J Appl Genet. 2023. PMID: 37380816
References
-
- Liu X., Zhou B., Yang H., Li Y., Yang Q., Lu Y., Gao Y. Sequencing and Analysis of Chrysanthemum carinatum Schousb and Kalimeris indica. The Complete Chloroplast Genomes Reveal Two Inversions and rbcL as Barcoding of the Vegetable. Molecules. 2018;23:1358. doi: 10.3390/molecules23061358. - DOI - PMC - PubMed
-
- Palmer J.D. The Molecular Biology of Plastids. Academic Press; Pittsburgh, PA, USA: 1991. CHAPTER 2-Plastid chromosomes: Structure and evolution.
-
- Jansen R.K., Raubeson L.A., Boore J.L., Depamphilis C.W., Chumley T.W., Haberle R.C., Wyman S.K., Alverson A.J., Peery R., Herman S.J., et al. Methods for Obtaining and Analyzing Whole Chloroplast Genome Sequences. Methods Enzymol. 2005;395:348–384. - PubMed
-
- Ma Y., Zhao L., Zhang W., Zhang Y., Xing X., Duan X., Hu J., Harris A.J., Liu P., Dai S., et al. Origins of cultivars of Chrysanthemum—Evidence from the chloroplast genome and nuclear LFY gene. J. Syst. Evol. 2020;58:925–944. doi: 10.1111/jse.12682. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

