Host Genetic Risk Factors Associated with COVID-19 Susceptibility and Severity in Vietnamese
- PMID: 36292769
- PMCID: PMC9601961
- DOI: 10.3390/genes13101884
Host Genetic Risk Factors Associated with COVID-19 Susceptibility and Severity in Vietnamese
Abstract
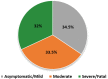
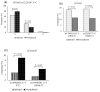
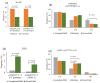
Since the emergence and rapid transmission of SARS-CoV-2, numerous scientific reports have searched for the association of host genetic variants with COVID-19, but the data are mostly acquired from Europe. In the current work, we explored the link between host genes (SARS-CoV-2 entry and immune system related to COVID-19 sensitivity/severity) and ABO blood types with COVID-19 from whole-exome data of 200 COVID-19 patients and 100 controls in Vietnam. The O blood type was found to be a protective factor that weakens the worst outcomes of infected individuals. For SARS-CoV-2 susceptibility, rs2229207 (TC genotype, allele C) and rs17860118 (allele T) of IFNAR2 increased the risk of infection, but rs139940581 (CT genotype, allele T) of SLC6A20 reduced virus sensitivity. For COVID-19 progress, the frequencies of rs4622692 (TG genotype) and rs1048610 (TC genotype) of ADAM17 were significantly higher in the moderate group than in the severe/fatal group. The variant rs12329760 (AA genotype) of TMPRSS2 was significantly associated with asymptomatic/mild symptoms. Additionally, rs2304255 (CT genotype, allele T) of TYK2 and rs2277735 (AG genotype) of DPP9 were associated with severe/fatal outcomes. Studies on different populations will give better insights into the pathogenesis, which is ethnic-dependent, and thus decipher the genetic factor's contribution to mechanisms that predispose people to being more vulnerable to COVID-19.
Keywords: ABO blood type; COVID-19; SARS-CoV-2; host genetic variants; whole-exome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Dadras O., Afsahi A.M., Pashaei Z., Mojdeganlou H., Karimi A., Habibi P., Barzegary A., Fakhfouri A., Mirzapour P., Janfaza N., et al. The Relationship between COVID-19 Viral Load and Disease Severity: A Systematic Review. Immun. Inflamm. Dis. 2022;10:e580. doi: 10.1002/iid3.580. - DOI - PMC - PubMed
-
- Vishnubhotla R., Vankadari N., Ketavarapu V., Amanchy R., Avanthi S., Bale G., Reddy D.N., Sasikala M. Genetic Variants in TMPRSS2 and Structure of SARS-CoV-2 Spike Glycoprotein and TMPRSS2 Complex. bioRxiv. 2020 doi: 10.1101/2020.06.30.179663. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

