A Novel De Novo NFKBIA Missense Mutation Associated to Ectodermal Dysplasia with Dysgammaglobulinemia
- PMID: 36292785
- PMCID: PMC9602067
- DOI: 10.3390/genes13101900
A Novel De Novo NFKBIA Missense Mutation Associated to Ectodermal Dysplasia with Dysgammaglobulinemia
Abstract
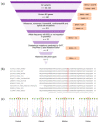
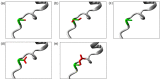
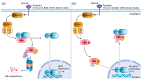
Background: Inborn errors of immunity (IEIs) are comprised of heterogeneous groups of genetic disorders affecting immune function. In this report, a 17-month-old Malay patient suspected of having Hyper IgM syndrome, a type of IEIs, was described. However, the diagnosis of Hyper IgM syndrome was excluded by the normal functional studies and the mild features of ectodermal dysplasia observed from a further clinical phenotype inspection. Methods: Whole-exome sequencing (WES) was performed to unravel the causative mutation in this patient. Results: The variant analysis demonstrated a novel missense mutation in NFKBIA (NM_020529:c.94A > T,NP_065390:p.Ser32Cys) and was predicted as damaging by in silico prediction tools. The NFKBIA gene encodes for IκBα, a member of nuclear factor kappa B (NF-κB) inhibitors, playing an important role in regulating NF-κB activity. The mutation occurred at the six degrons (Asp31-Ser36) in IκBα which were evolutionarily conserved across several species. Prediction analysis suggested that the substitution of Ser32Cys may cause a loss of the phosphorylation site at residue 32 and a gain of the sumoylation site at residue 38, resulting in the alteration of post-translational modifications of IκBα required for NF-κB activation. Conclusion: Our analysis hints that the post-translational modification in the NFKBIA Ser32Cys mutant would alter the signaling pathway of NF-κB. Our findings support the usefulness of WES in diagnosing IEIs and suggest the role of post-translational modification of IκBα.
Keywords: IκBα; NF-κB; NFKBIA; hyper IgM-like phenotype; post-translational modification.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Heterozygous N-terminal deletion of IkappaBalpha results in functional nuclear factor kappaB haploinsufficiency, ectodermal dysplasia, and immune deficiency.J Allergy Clin Immunol. 2007 Oct;120(4):900-7. doi: 10.1016/j.jaci.2007.08.035. J Allergy Clin Immunol. 2007. PMID: 17931563
-
Autosomal dominant anhidrotic ectodermal dysplasia with immunodeficiency caused by a novel NFKBIA mutation, p.Ser36Tyr, presents with mild ectodermal dysplasia and non-infectious systemic inflammation.J Clin Immunol. 2013 Oct;33(7):1165-74. doi: 10.1007/s10875-013-9924-z. Epub 2013 Jul 18. J Clin Immunol. 2013. PMID: 23864385
-
A novel mutation in NFKBIA/IKBA results in a degradation-resistant N-truncated protein and is associated with ectodermal dysplasia with immunodeficiency.Hum Mutat. 2008 Jun;29(6):861-8. doi: 10.1002/humu.20740. Hum Mutat. 2008. PMID: 18412279 Free PMC article.
-
Diagnosis and treatment in anhidrotic ectodermal dysplasia with immunodeficiency.Allergol Int. 2012 Jun;61(2):207-17. doi: 10.2332/allergolint.12-RAI-0446. Allergol Int. 2012. PMID: 22635013 Review.
-
Human IκBα Gain of Function: a Severe and Syndromic Immunodeficiency.J Clin Immunol. 2017 Jul;37(5):397-412. doi: 10.1007/s10875-017-0400-z. Epub 2017 Jun 9. J Clin Immunol. 2017. PMID: 28597146 Free PMC article. Review.
Cited by
-
Genetic Comparison and Selection of Reproductive and Growth-Related Traits in Qinchuan Cattle and Two Belgian Cattle Breeds.Animals (Basel). 2025 Feb 19;15(4):608. doi: 10.3390/ani15040608. Animals (Basel). 2025. PMID: 40003088 Free PMC article.
-
The Revolution of Genetic Diagnosis: An Example from Rare Disorders.Genes (Basel). 2024 Oct 15;15(10):1328. doi: 10.3390/genes15101328. Genes (Basel). 2024. PMID: 39457452 Free PMC article.
-
Compound heterozygous WNT10A missense variations exacerbated the tooth agenesis caused by hypohidrotic ectodermal dysplasia.BMC Oral Health. 2024 Jan 27;24(1):136. doi: 10.1186/s12903-024-03888-5. BMC Oral Health. 2024. PMID: 38280992 Free PMC article.
References
-
- Bousfiha A., Jeddane L., Picard C., Al-Herz W., Ailal F., Chatila T., Cunningham-Rundles C., Etzioni A., Franco J.L., Holland S.M., et al. Human Inborn Errors of Immunity: 2019 Update of the IUIS Phenotypical Classification. J. Clin. Immunol. 2020;40:66–81. doi: 10.1007/s10875-020-00758-x. - DOI - PMC - PubMed
-
- Tangye S.G., Al-Herz W., Bousfiha A., Cunningham-Rundles C., Franco J.L., Holland S.M., Klein C., Morio T., Oksenhendler E., Picard C., et al. Human Inborn Errors of Immunity: 2022 Update on the Classification from the International Union of Immunological Societies Expert Committee. J. Clin. Immunol. 2022:1–35. doi: 10.1007/s10875-022-01289-3. - DOI - PMC - PubMed
-
- Engelbrecht C., Urban M., Schoeman M., Paarwater B., van Coller A., Abraham D.R., Cornelissen H., Glashoff R., Esser M., Möller M., et al. Clinical Utility of Whole Exome Sequencing and Targeted Panels for the Identification of Inborn Errors of Immunity in a Resource-Constrained Setting. Front. Immunol. 2021;12:665621. doi: 10.3389/fimmu.2021.665621. - DOI - PMC - PubMed
-
- Ripen A.M., Chear C.T., Baharin M.F., Nallusamy R., Chan K.C., Kassim A., Choo C.M., Wong K.J., Fong S.M., Tan K.K., et al. A Single-center Pilot Study in Malaysia on the Clinical Utility of Whole-exome Sequencing for Inborn Errors of Immunity. Clin. Exp. Immunol. 2021;206:119–128. doi: 10.1111/cei.13626. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

