Genome-Wide Identification of Potential mRNAs in Drought Response in Wheat (Triticum aestivum L.)
- PMID: 36292791
- PMCID: PMC9601369
- DOI: 10.3390/genes13101906
Genome-Wide Identification of Potential mRNAs in Drought Response in Wheat (Triticum aestivum L.)
Abstract
Plant cell metabolism inevitably forms an important drought-responsive mechanism, which halts crop productivity. Globally, more than 30% of the total harvested area was affected by dehydration. RNA-seq technology has enabled biologists to identify stress-responsive genes in relatively quick times. However, one shortcoming of this technology is the inconsistent data generation compared to other parts of the world. So, we have tried, here, to generate a consensus by analyzing meta-transcriptomic data available in the public microarray database GEO NCBI. In this way, the aim was set, here, to identify stress genes commonly identified as differentially expressed (p < 0.05) then followed by downstream analyses. The search term “Drought in wheat” resulted in 233 microarray experiments from the GEO NCBI database. After discarding empty datasets containing no expression data, the large-scale meta-transcriptome analytics and one sample proportional test were carried out (Bonferroni adjusted p < 0.05) to reveal a set of 11 drought-responsive genes on a global scale. The annotation of these genes revealed that the transcription factor activity of RNA polymerase II and sequence-specific DNA-binding mechanism had a significant role during the drought response in wheat. Similarly, the primary root differentiation zone annotations, controlled by TraesCS5A02G456300 and TraesCS7B02G243600 genes, were found as top-enriched terms (p < 0.05 and Q < 0.05). The resultant standard drought genes, glycosyltransferase; Arabidopsis thaliana KNOTTED-like; bHLH family protein; Probable helicase MAGATAMA 3; SBP family protein; Cytochrome c oxidase subunit 2; Trihelix family protein; Mic1 domain-containing protein; ERF family protein; HD-ZIP I protein; and ERF family protein, are important in terms of their worldwide proved link with stress. From a future perspective, this study could be important in a breeding program contributing to increased crop yield. Moreover, the wheat varieties could be identified as drought-resistant/sensitive based on the nature of gene expression levels.
Keywords: RNA seq; bread wheat; drought; genomics; meta data.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
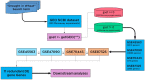





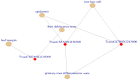
References
-
- Zulkiffal M., Ahsan A., Ahmed J., Musa M., Kanwal A., Saleem M., Anwar J., ur Rehman A., Ajmal S., Gulnaz S., et al. Heat and Drought Stresses in Wheat (Triticum aestivum L.): Substantial Yield Losses, Practical Achievements, Improvement Approaches, and Adaptive Mechanisms. Plant Stress Physiol. 2021;3 doi: 10.5772/intechopen.92378. - DOI
-
- Hashmi H.A., Belgacem A.O., Behnassi M., Javed K., Baig M.B. Emerging Challenges to Food Production and Security in Asia, Middle East, and Africa. Springer; Cham, Switzherland: 2021. Impacts of Climate Change on Livestock and Related Food Security Implications—Overview of the Situation in Pakistan and Policy Recommendations; pp. 197–239. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

