LncGSAR Controls Ovarian Granulosa Cell Steroidogenesis via Sponging MiR-125b to Activate SCAP/SREBP Pathway
- PMID: 36293007
- PMCID: PMC9603659
- DOI: 10.3390/ijms232012132
LncGSAR Controls Ovarian Granulosa Cell Steroidogenesis via Sponging MiR-125b to Activate SCAP/SREBP Pathway
Abstract
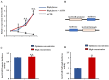
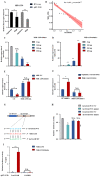
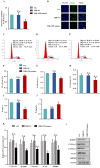
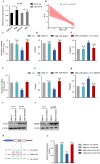
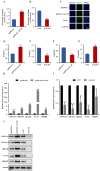
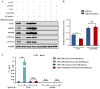
Long non-coding RNAs (lncRNAs) have been shown to play important roles in livestock fecundity, and many lncRNAs that affect follicular development and reproductive diseases have been identified in the ovary. However, only a few of them have been functionally annotated and mechanistically validated. In this study, we identified a new lncRNA (lncGSAR) and investigated its effects on the proliferation and steroidogenesis of ovine granulosa cells (GCs). High concentrations of glucose (add 33.6 mM glucose) caused high expression of lncGSAR in GCs by regulating its stability, and lncGSAR overexpression promoted GCs proliferation, estrogen secretion, and inhibited progesterone secretion, whereas interference with lncGASR had the opposite effect. Next, we found that the RNA molecules of lncGSAR act on MiR-125b as competitive endogenous RNA (ceRNA), and SREBP-cleavage-activating protein (SCAP) was verified as a target of MiR-125b. LncGASR overexpression increased the expression of SCAP, SREBP, and steroid hormone-related proteins, which can be attenuated by MiR-125b. Our results demonstrated that lncGSAR can act as a ceRNA to activate SCAP/SREBP signaling by sponging MiR-125b to regulate steroid hormone secretion in GCs. These findings provide new insights into the mechanisms of nutrient-regulated follicle development in ewes.
Keywords: MiR-125b; ceRNA; granulosa cell; lncGSAR; steroidogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
LncRNA SNHG12 promotes cell proliferation and inhibits apoptosis of granulosa cells in polycystic ovarian syndrome by sponging miR-129 and miR-125b.J Ovarian Res. 2024 Apr 2;17(1):72. doi: 10.1186/s13048-024-01392-6. J Ovarian Res. 2024. PMID: 38566229 Free PMC article.
-
Long non-coding RNA TCONS_00814106 regulates porcine granulosa cell proliferation and apoptosis by sponging miR-1343.Mol Cell Endocrinol. 2021 Jan 15;520:111064. doi: 10.1016/j.mce.2020.111064. Epub 2020 Oct 19. Mol Cell Endocrinol. 2021. PMID: 33091558
-
LncRNA-412.25 activates the LIF/STAT3 signaling pathway in ovarian granulosa cells of Hu sheep by sponging miR-346.FASEB J. 2022 Sep;36(9):e22467. doi: 10.1096/fj.202200632R. FASEB J. 2022. PMID: 35929417
-
Long non-coding RNA MSTRG.5970.28 regulates proliferation and apoptosis of goose follicle granulosa cells via the miR-133a-3p/ANOS1 pathway.Poult Sci. 2023 Mar;102(3):102451. doi: 10.1016/j.psj.2022.102451. Epub 2022 Dec 25. Poult Sci. 2023. PMID: 36634463 Free PMC article.
-
miR-125b Contributes to Ovarian Granulosa Cell Apoptosis Through Targeting BMPR1B, a Major Gene for Sheep Prolificacy.Reprod Sci. 2019 Feb;26(2):295-305. doi: 10.1177/1933719118770544. Epub 2018 Apr 16. Reprod Sci. 2019. PMID: 29661099
Cited by
-
Design and verification of a 25 K multiple-SNP liquid-capture chip by target sequencing for dairy goat.BMC Genomics. 2025 Apr 15;26(1):377. doi: 10.1186/s12864-025-11576-z. BMC Genomics. 2025. PMID: 40234771 Free PMC article.
-
TGF-β1 Mediates Novel-m0297-5p Targeting WNT5A to Participate in the Proliferation of Ovarian Granulosa Cells in Small-Tailed Han Sheep.Int J Mol Sci. 2025 Feb 24;26(5):1961. doi: 10.3390/ijms26051961. Int J Mol Sci. 2025. PMID: 40076587 Free PMC article.
-
Lnc MSTRG 4701.7 targets miR-1786/RORa to competitively regulate proliferation and apoptosis in chicken follicular granulosa cells.Front Vet Sci. 2025 Apr 30;12:1583287. doi: 10.3389/fvets.2025.1583287. eCollection 2025. Front Vet Sci. 2025. PMID: 40370832 Free PMC article.
-
Long non-coding RNA Loc105611671 promotes the proliferation of ovarian granulosa cells and steroid hormone production upregulation of CDC42.Front Vet Sci. 2024 Mar 4;11:1366759. doi: 10.3389/fvets.2024.1366759. eCollection 2024. Front Vet Sci. 2024. PMID: 38500606 Free PMC article.
-
Prolactin Inhibition Promotes Follicle Recruitment by Increasing PIKfyve Expression in Ewes During the Estrus Stage.Animals (Basel). 2024 Dec 7;14(23):3541. doi: 10.3390/ani14233541. Animals (Basel). 2024. PMID: 39682506 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

