Characterization of the Human Papillomavirus 16 Oncogenes in K14HPV16 Mice: Sublineage A1 Drives Multi-Organ Carcinogenesis
- PMID: 36293226
- PMCID: PMC9604181
- DOI: 10.3390/ijms232012371
Characterization of the Human Papillomavirus 16 Oncogenes in K14HPV16 Mice: Sublineage A1 Drives Multi-Organ Carcinogenesis
Abstract
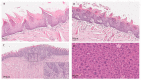
The study of human papillomavirus (HPV)-induced carcinogenesis uses multiple in vivo mouse models, one of which relies on the cytokeratin 14 gene promoter to drive the expression of all HPV early oncogenes. This study aimed to determine the HPV16 variant and sublineage present in the K14HPV16 mouse model. This information can be considered of great importance to further enhance this K14HPV16 model as an essential research tool and optimize its use for basic and translational studies. Our study evaluated HPV DNA from 17 samples isolated from 4 animals, both wild-type (n = 2) and HPV16-transgenic mice (n = 2). Total DNA was extracted from tissues and the detection of HPV16 was performed using a qPCR multiplex. HPV16-positive samples were subsequently whole-genome sequenced by next-generation sequencing techniques. The phylogenetic positioning clearly shows K14HPV16 samples clustering together in the sub-lineage A1 (NC001526.4). A comparative genome analysis of K14HPV16 samples revealed three mutations to the human papillomaviruses type 16 sublineage A1 representative strain. Knowledge of the HPV 16 variant is fundamental, and these findings will allow the rational use of this animal model to explore the role of the A1 sublineage in HPV-driven cancer.
Keywords: HPV16; K14HPV16; carcinogenesis; lineage; variant.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

