Truncated Analogues of a G-Quadruplex-Forming Aptamer Targeting Mutant Huntingtin: Shorter Is Better!
- PMID: 36293267
- PMCID: PMC9604342
- DOI: 10.3390/ijms232012412
Truncated Analogues of a G-Quadruplex-Forming Aptamer Targeting Mutant Huntingtin: Shorter Is Better!
Abstract
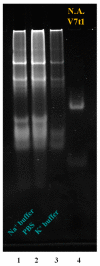
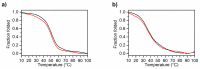
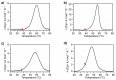
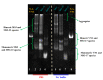
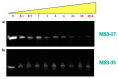
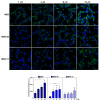
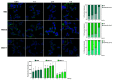
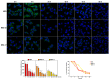
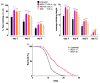
Two analogues of the MS3 aptamer, which was previously shown to have an exquisite capability to selectively bind and modulate the activity of mutant huntingtin (mHTT), have been here designed and evaluated in their physicochemical and biological properties. Featured by a distinctive propensity to form complex G-quadruplex structures, including large multimeric aggregates, the original 36-mer MS3 has been truncated to give a 33-mer (here named MS3-33) and a 17-mer (here named MS3-17). A combined use of different techniques (UV, CD, DSC, gel electrophoresis) allowed a detailed physicochemical characterization of these novel G-quadruplex-forming aptamers, tested in vitro on SH-SY5Y cells and in vivo on a Drosophila Huntington's disease model, in which these shorter MS3-derived oligonucleotides proved to have improved bioactivity in comparison with the parent aptamer.
Keywords: Drosophila melanogaster model; G-quadruplex; Huntington’s disease; aptamers; physicochemical characterization.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

