The OsCBL8-OsCIPK17 Module Regulates Seedling Growth and Confers Resistance to Heat and Drought in Rice
- PMID: 36293306
- PMCID: PMC9604039
- DOI: 10.3390/ijms232012451
The OsCBL8-OsCIPK17 Module Regulates Seedling Growth and Confers Resistance to Heat and Drought in Rice
Abstract
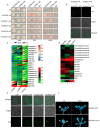
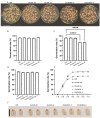
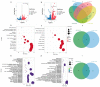
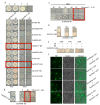
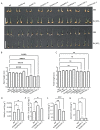
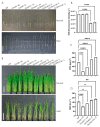
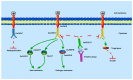
The calcium signaling pathway is critical for plant growth, development, and response to external stimuli. The CBL-CIPK pathway has been well characterized as a calcium-signaling pathway. However, in most reports, only a single function for this module has been described. Here, we examined multiple functions of this module. CIPK showed a similar distribution to that of CBL, and OsCBL and OsCIPK families were retained after experiencing whole genome duplication events through the phylogenetic and synteny analysis. This study found that OsCBL8 negatively regulated rice seed germination and seedling growth by interacting with OsCIPK17 with overexpression and gene editing mutant plants as materials combining plant phenotype, physiological indicators and transcriptome sequencing. This process is likely mediated by OsPP2C77, which is a member of the ABA signaling pathway. In addition, OsCBL mediated the targeting of OsNAC77 and OsJAMYB by OsCIPK17, thus conferring resistance to high temperatures and pathogens in rice. Our work reveals a unique signaling pathway, wherein OsCBL8 interacts with OsCIPK17 and provides rice with multiple resistance while also regulating seedling growth.
Keywords: CBL; CIPK; Oryza sativa; drought; growth; high temperature.
Conflict of interest statement
The authors have no conflict of interest.
Figures


Similar articles
-
Combined metabolomic and transcriptomic analysis reveals key components of OsCIPK17 overexpression improves drought tolerance in rice.Front Plant Sci. 2023 Jan 9;13:1043757. doi: 10.3389/fpls.2022.1043757. eCollection 2022. Front Plant Sci. 2023. PMID: 36699859 Free PMC article.
-
Comprehensive structural, interaction and expression analysis of CBL and CIPK complement during abiotic stresses and development in rice.Cell Calcium. 2014 Aug;56(2):81-95. doi: 10.1016/j.ceca.2014.05.003. Epub 2014 Jun 5. Cell Calcium. 2014. PMID: 24970010
-
Overexpression of a new stress-repressive gene OsDSR2 encoding a protein with a DUF966 domain increases salt and simulated drought stress sensitivities and reduces ABA sensitivity in rice.Plant Cell Rep. 2014 Feb;33(2):323-36. doi: 10.1007/s00299-013-1532-0. Epub 2013 Nov 20. Plant Cell Rep. 2014. PMID: 24247850
-
Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance.BMC Plant Biol. 2016 Sep 20;16(1):203. doi: 10.1186/s12870-016-0897-y. BMC Plant Biol. 2016. PMID: 27646344 Free PMC article.
-
Wheat CBL-interacting protein kinase 23 positively regulates drought stress and ABA responses.BMC Plant Biol. 2018 May 25;18(1):93. doi: 10.1186/s12870-018-1306-5. BMC Plant Biol. 2018. PMID: 29801463 Free PMC article.
Cited by
-
Combined metabolomic and transcriptomic analysis reveals key components of OsCIPK17 overexpression improves drought tolerance in rice.Front Plant Sci. 2023 Jan 9;13:1043757. doi: 10.3389/fpls.2022.1043757. eCollection 2022. Front Plant Sci. 2023. PMID: 36699859 Free PMC article.
-
Evolution and subfunctionalization of CIPK6 homologous genes in regulating cotton drought resistance.Nat Commun. 2024 Jul 9;15(1):5733. doi: 10.1038/s41467-024-50097-3. Nat Commun. 2024. PMID: 38977687 Free PMC article.
-
Calcineurin B-like protein ZmCBL8-1 promotes salt stress resistance in Arabidopsis.Planta. 2024 Jan 29;259(2):49. doi: 10.1007/s00425-024-04330-4. Planta. 2024. PMID: 38285217
-
Identification and expression pattern analysis of the OsCBL gene family in rice.Front Plant Sci. 2025 Jul 9;16:1625014. doi: 10.3389/fpls.2025.1625014. eCollection 2025. Front Plant Sci. 2025. PMID: 40703858 Free PMC article.
-
Calcium Signaling and the Response to Heat Shock in Crop Plants.Int J Mol Sci. 2023 Dec 26;25(1):324. doi: 10.3390/ijms25010324. Int J Mol Sci. 2023. PMID: 38203495 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

