Identification and Investigation of the Genetic Variations and Candidate Genes Responsible for Seed Weight via GWAS in Paper Mulberry
- PMID: 36293375
- PMCID: PMC9604540
- DOI: 10.3390/ijms232012520
Identification and Investigation of the Genetic Variations and Candidate Genes Responsible for Seed Weight via GWAS in Paper Mulberry
Abstract
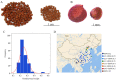
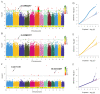
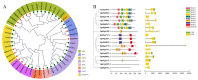
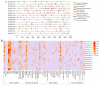
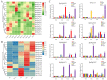
Seeds directly determine the survival and population size of woody plants, but the genetic basis of seed weight in woody plants remain poorly explored. To identify genetic variations and candidate genes responsible for seed weight in natural woody populations, we investigated the hundred-seed weight of 198 paper mulberry individuals from different areas. Our results showed that the hundred-seed weight of paper mulberry was significantly associated with the bioclimatic variables of sampling sites, which increased from south to north along the latitudinal-temperature gradient. Using 2,414,978 high-quality SNPs from re-sequencing data, the genome-wide association analysis of the hundred-seed weight was performed under three models, which identified 148, 19 and 12 associated genes, respectively. Among them, 25 candidate genes were directly hit by the significant SNPs, including the WRKY transcription factor, fatty acid desaturase, F-box protein, etc. Most importantly, we identified three crucial genetic variations in the coding regions of candidate genes (Bp02g2123, Bp01g3291 and Bp10g1642), and significant differences in the hundred-seed weight were detected among the individuals carrying different genotypes. Further analysis revealed that Bp02g2123 encoding a fatty acid desaturase (FAD) might be a key factor affecting the seed weight and local climate adaptation of woody plants. Furthermore, the genome-wide investigation and expression analysis of FAD genes were performed, and the results suggested that BpFADs widely expressed in various tissues and responded to multiple phytohormone and stress treatments. Overall, our study identifies valuable genetic variations and candidate genes, and provides a better understanding of the genetic basis of seed weight in woody plants.
Keywords: candidate gene; fatty acid desaturase; genetic variation; genome-wide association study; seed weight; woody plant.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
Natural population re-sequencing detects the genetic basis of local adaptation to low temperature in a woody plant.Plant Mol Biol. 2021 Apr;105(6):585-599. doi: 10.1007/s11103-020-01111-x. Epub 2021 Mar 2. Plant Mol Biol. 2021. PMID: 33651261
-
Genome-wide association study and transcriptome analysis reveal the genetic basis underlying the environmental adaptation of plant height in a woody plant.Plant Physiol Biochem. 2025 Feb;219:109361. doi: 10.1016/j.plaphy.2024.109361. Epub 2024 Nov 28. Plant Physiol Biochem. 2025. PMID: 39631345
-
Genome-Wide Association Study in Arabidopsis thaliana of Natural Variation in Seed Oil Melting Point: A Widespread Adaptive Trait in Plants.J Hered. 2016 May;107(3):257-65. doi: 10.1093/jhered/esw008. Epub 2016 Feb 10. J Hered. 2016. PMID: 26865732 Free PMC article.
-
In Silico Analysis of Fatty Acid Desaturases Structures in Camelina sativa, and Functional Evaluation of Csafad7 and Csafad8 on Seed Oil Formation and Seed Morphology.Int J Mol Sci. 2021 Oct 8;22(19):10857. doi: 10.3390/ijms221910857. Int J Mol Sci. 2021. PMID: 34639198 Free PMC article.
-
Response Mechanisms of Woody Plants to High-Temperature Stress.Plants (Basel). 2023 Oct 22;12(20):3643. doi: 10.3390/plants12203643. Plants (Basel). 2023. PMID: 37896106 Free PMC article. Review.
Cited by
-
Integrating genome-wide association and transcriptome analysis to provide molecular insights into heterophylly and eco-adaptability in woody plants.Hortic Res. 2023 Nov 17;10(11):uhad212. doi: 10.1093/hr/uhad212. eCollection 2023 Nov. Hortic Res. 2023. PMID: 38046852 Free PMC article.
References
-
- Lu Z., Zhao S., Gang C. Relationships between chemical compositions of quercus species seeds and climatic factors in temperate zone of NSTEC. Acta Ecol. Sin. 2012;32:7857–7865.
-
- Harel D., Holzapfel C., Sternberg M. Seed mass and dormancy of annual plant populations and communities decreases with aridity and rainfall predictability. Basic Appl. Ecol. 2011;12:674–684. doi: 10.1016/j.baae.2011.09.003. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

