Drug Repurposing for Cystic Fibrosis: Identification of Drugs That Induce CFTR-Independent Fluid Secretion in Nasal Organoids
- PMID: 36293514
- PMCID: PMC9603984
- DOI: 10.3390/ijms232012657
Drug Repurposing for Cystic Fibrosis: Identification of Drugs That Induce CFTR-Independent Fluid Secretion in Nasal Organoids
Abstract
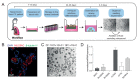
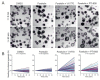
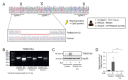
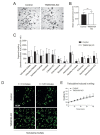
Individuals with cystic fibrosis (CF) suffer from severe respiratory disease due to a genetic defect in the cystic fibrosis transmembrane conductance regulator (CFTR) gene, which impairs airway epithelial ion and fluid secretion. New CFTR modulators that restore mutant CFTR function have been recently approved for a large group of people with CF (pwCF), but ~19% of pwCF cannot benefit from CFTR modulators Restoration of epithelial fluid secretion through non-CFTR pathways might be an effective treatment for all pwCF. Here, we developed a medium-throughput 384-well screening assay using nasal CF airway epithelial organoids, with the aim to repurpose FDA-approved drugs as modulators of non-CFTR-dependent epithelial fluid secretion. From a ~1400 FDA-approved drug library, we identified and validated 12 FDA-approved drugs that induced CFTR-independent fluid secretion. Among the hits were several cAMP-mediating drugs, including β2-adrenergic agonists. The hits displayed no effects on chloride conductance measured in the Ussing chamber, and fluid secretion was not affected by TMEM16A, as demonstrated by knockout (KO) experiments in primary nasal epithelial cells. Altogether, our results demonstrate the use of primary nasal airway cells for medium-scale drug screening, target validation with a highly efficient protocol for generating CRISPR-Cas9 KO cells and identification of compounds which induce fluid secretion in a CFTR- and TMEM16A-indepent manner.
Keywords: TMEM16A; cystic fibrosis; drug repurposing; nasal organoids; screening assay.
Conflict of interest statement
J.M.B. has a patent granted (10006904) related to CFTR function measurements in organoids and received personal fees from HUB/Royal Dutch academy of sciences, during the conduct of the study; nonfinancial support from Vertex Pharmaceuticals and personal fees and nonfinancial support from Proteostasis Therapeutics, outside the submitted work. C.K.v.d.E. reports grants from GSK, Nutricia, TEVA, Gilead, Vertex, ProQR, Proteostasis, Galapagos NV, Eloxx pharmaceuticals, outside the submitted work; In addition, C.K.v.d.E. has a patent related to CFTR function measurements in organoids (10006904) with royalties paid.
Figures








References
-
- Riordan J.R., Rommens J.M., Kerem B.S., Alon N.O.A., Rozmahel R., Grzelczak Z., Zielenski J., Lok S.I., Plavsic N., Chou J.L., et al. Identification of the Cystic Fibrosis Gene: Cloning and Characterization of Complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. - DOI - PubMed
-
- Middleton P.G., Mall M.A., Dřevínek P., Lands L.C., McKone E.F., Polineni D., Ramsey B.W., Taylor-Cousar J.L., Tullis E., Vermeulen F., et al. Elexacaftor-Tezacaftor-Ivacaftor for Cystic Fibrosis with a Single Phe508del Allele. N. Engl. J. Med. 2019;381:1809–1819. doi: 10.1056/NEJMoa1908639. - DOI - PMC - PubMed
-
- Heijerman H.G.M., McKone E.F., Downey D.G., Van Braeckel E., Rowe S.M., Tullis E., Mall M.A., Welter J.J., Ramsey B.W., McKee C.M., et al. Efficacy and Safety of the Elexacaftor plus Tezacaftor plus Ivacaftor Combination Regimen in People with Cystic Fibrosis Homozygous for the F508del Mutation: A Double-Blind, Randomised, Phase 3 Trial. Lancet. 2019;394:1940–1948. doi: 10.1016/S0140-6736(19)32597-8. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

