The Transcription Factor CsAtf1 Negatively Regulates the Cytochrome P450 Gene CsCyp51G1 to Increase Fludioxonil Sensitivity in Colletotrichum siamense
- PMID: 36294597
- PMCID: PMC9605597
- DOI: 10.3390/jof8101032
The Transcription Factor CsAtf1 Negatively Regulates the Cytochrome P450 Gene CsCyp51G1 to Increase Fludioxonil Sensitivity in Colletotrichum siamense
Abstract
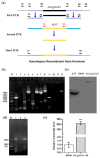
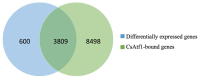
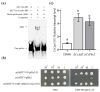
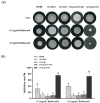
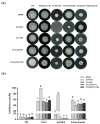
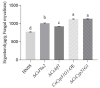
Previous studies have shown that the high-osmolarity glycerol mitogen-activated protein kinase (HOG MAPK) signaling pathway and its downstream transcription factor CsAtf1 are involved in the regulation of fludioxonil sensitivity in C. siamense. However, the downstream target genes of CsAtf1 related to the fludioxonil stress response remain unclear. Here, we performed chromatin immunoprecipitation sequencing (ChIP-Seq) and high-throughput RNA-sequencing (RNA-Seq) to identify genome-wide potential CsAtf1 target genes. A total of 3809 significantly differentially expressed genes were predicted to be directly regulated by CsAtf1, including 24 cytochrome oxidase-related genes. Among them, a cytochrome P450-encoding gene, designated CsCyp51G1, was confirmed to be a target gene, and its transcriptional expression was negatively regulated by CsAtf1, as determined using an electrophoretic mobility shift assay (EMSA), a yeast one-hybrid (Y1H) assay, and quantitative real-time PCR (qRT-PCR). Moreover, the overexpression mutant CsCYP51G1 of C. siamense exhibited increased fludioxonil tolerance, and the CsCYP51G1 deletion mutant exhibited decreased fludioxonil resistance, which revealed that CsCyp51G1 is involved in fludioxonil sensitivity regulation in C. siamense. However, the cellular ergosterol content of the mutants was not consistent with the phenotype of fludioxonil sensitivity, which indicated that CsCyp51G1 regulates fludioxonil sensitivity by affecting factors other than the ergosterol level in C. siamense. In conclusion, our data indicate that the transcription factor CsAtf1 negatively regulates the cytochrome P450 gene CsCyp51G1 to increase fludioxonil sensitivity in C. siamense.
Keywords: ChIP-Seq; Colletotrichum siamense; CsAtf1; CsCyp51G1; RNA-Seq; cytochrome P450; fludioxonil sensitivity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- O’Connell R.J., Thon M.R., Hacquard S., Amyotte S.G., Kleemann J., Torres M., Damm U., Buiate E.A., Epstein L., Alkan N., et al. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat. Genet. 2012;44:1060–1065. doi: 10.1038/ng.2372. - DOI - PMC - PubMed
-
- Qin L.P., Zhang Y., Su Q., Chen Y.L., Nong Q., Xie L., Yu G.M., Huang S.L. First report of anthracnose of Mangifera indica caused by Colletotrichum scovillei in China. Plant Dis. 2019;103:1043. doi: 10.1094/PDIS-11-18-1980-PDN. - DOI
-
- Huang R., Sun W., Wang L., Li Q., Huang S., Tang L., Guo T., Mo J., Hsiang T. Identification and characterization of Colletotrichum species associated with anthracnose disease of banana. Plant Pathol. 2021;70:1827–1837. doi: 10.1111/ppa.13426. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

