Identification of Potential Repurposable Drugs in Alzheimer's Disease Exploiting a Bioinformatics Analysis
- PMID: 36294870
- PMCID: PMC9605472
- DOI: 10.3390/jpm12101731
Identification of Potential Repurposable Drugs in Alzheimer's Disease Exploiting a Bioinformatics Analysis
Abstract
Alzheimer's disease (AD) is a neurologic disorder causing brain atrophy and the death of brain cells. It is a progressive condition marked by cognitive and behavioral impairment that significantly interferes with daily activities. AD symptoms develop gradually over many years and eventually become more severe, and no cure has been found yet to arrest this process. The present study is directed towards suggesting putative novel solutions and paradigms for fighting AD pathogenesis by exploiting new insights from network medicine and drug repurposing strategies. To identify new drug-AD associations, we exploited SAveRUNNER, a recently developed network-based algorithm for drug repurposing, which quantifies the vicinity of disease-associated genes to drug targets in the human interactome. We complemented the analysis with an in silico validation of the candidate compounds through a gene set enrichment analysis, aiming to determine if the modulation of the gene expression induced by the predicted drugs could be counteracted by the modulation elicited by the disease. We identified some interesting compounds belonging to the beta-blocker family, originally approved for treating hypertension, such as betaxolol, bisoprolol, and metoprolol, whose connection with a lower risk to develop Alzheimer's disease has already been observed. Moreover, our algorithm predicted multi-kinase inhibitors such as regorafenib, whose beneficial effects were recently investigated for neuroinflammation and AD pathology, and mTOR inhibitors such as sirolimus, whose modulation has been associated with AD.
Keywords: dementia; drug repurposing; network theory.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
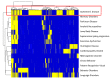

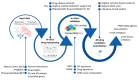
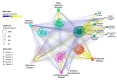
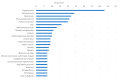
References
LinkOut - more resources
Full Text Sources
Miscellaneous

