Intronization Signatures in Coding Exons Reveal the Evolutionary Fluidity of Eukaryotic Gene Architecture
- PMID: 36296178
- PMCID: PMC9612004
- DOI: 10.3390/microorganisms10101901
Intronization Signatures in Coding Exons Reveal the Evolutionary Fluidity of Eukaryotic Gene Architecture
Abstract
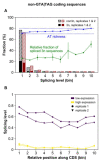
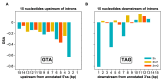
The conventionally clear distinction between exons and introns in eukaryotic genes is actually blurred. To illustrate this point, consider sequences that are retained in mature mRNAs about 50% of the time: how should they be classified? Moreover, although it is clear that RNA splicing influences gene expression levels and is an integral part of interdependent cellular networks, introns continue to be regarded as accidental insertions; exogenous sequences whose evolutionary origin is independent of mRNA-associated processes and somewhat still elusive. Here, we present evidence that aids to resolve this disconnect between conventional views about introns and current knowledge about the role of RNA splicing in the eukaryotic cell. We first show that coding sequences flanked by cryptic splice sites are negatively selected on a genome-wide scale in Paramecium. Then, we exploit selection intensity to infer splicing-related evolutionary dynamics. Our analyses suggest that intron gain begins as a splicing error, involves a transient phase of alternative splicing, and is preferentially completed at the 5' end of genes, which through intron gain can become highly expressed. We conclude that relaxed selective constraints may promote biological complexity in Paramecium and that the relationship between exons and introns is fluid on an evolutionary scale.
Keywords: RNA splicing; alternative splicing; exon; exonization; gene architecture; gene expression; intron; intronization; purifying selection.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
Similar articles
-
De Novo Creation of Two Novel Spliceosomal Introns of RECG1 by Intronization of Formerly Exonic Sequences in Orchidaceae.J Mol Evol. 2025 Apr;93(2):267-277. doi: 10.1007/s00239-025-10242-y. Epub 2025 Apr 9. J Mol Evol. 2025. PMID: 40202594
-
The architecture of pre-mRNAs affects mechanisms of splice-site pairing.Proc Natl Acad Sci U S A. 2005 Nov 8;102(45):16176-81. doi: 10.1073/pnas.0508489102. Epub 2005 Oct 31. Proc Natl Acad Sci U S A. 2005. PMID: 16260721 Free PMC article.
-
Origin and evolution of spliceosomal introns.Biol Direct. 2012 Apr 16;7:11. doi: 10.1186/1745-6150-7-11. Biol Direct. 2012. PMID: 22507701 Free PMC article. Review.
-
Changes in exon-intron structure during vertebrate evolution affect the splicing pattern of exons.Genome Res. 2012 Jan;22(1):35-50. doi: 10.1101/gr.119834.110. Epub 2011 Oct 5. Genome Res. 2012. PMID: 21974994 Free PMC article.
-
Exonization of transposed elements: A challenge and opportunity for evolution.Biochimie. 2011 Nov;93(11):1928-34. doi: 10.1016/j.biochi.2011.07.014. Epub 2011 Jul 26. Biochimie. 2011. PMID: 21787833 Review.
Cited by
-
De Novo Creation of Two Novel Spliceosomal Introns of RECG1 by Intronization of Formerly Exonic Sequences in Orchidaceae.J Mol Evol. 2025 Apr;93(2):267-277. doi: 10.1007/s00239-025-10242-y. Epub 2025 Apr 9. J Mol Evol. 2025. PMID: 40202594
-
Discovering Intron Gain Events in Humans Through Large-Scale Evolutionary Comparisons.Genome Biol Evol. 2025 May 30;17(6):evaf091. doi: 10.1093/gbe/evaf091. Genome Biol Evol. 2025. PMID: 40380889 Free PMC article.
-
A Review for the Special Issue on Paramecium as a Modern Model Organism.Microorganisms. 2023 Apr 3;11(4):937. doi: 10.3390/microorganisms11040937. Microorganisms. 2023. PMID: 37110360 Free PMC article. Review.
-
Introns: the "dark matter" of the eukaryotic genome.Front Genet. 2023 May 16;14:1150212. doi: 10.3389/fgene.2023.1150212. eCollection 2023. Front Genet. 2023. PMID: 37260773 Free PMC article. Review.
-
Paramecium Genetics, Genomics, and Evolution.Annu Rev Genet. 2023 Nov 27;57:391-410. doi: 10.1146/annurev-genet-071819-104035. Annu Rev Genet. 2023. PMID: 38012024 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources

