Intronization Signatures in Coding Exons Reveal the Evolutionary Fluidity of Eukaryotic Gene Architecture
- PMID: 36296178
- PMCID: PMC9612004
- DOI: 10.3390/microorganisms10101901
Intronization Signatures in Coding Exons Reveal the Evolutionary Fluidity of Eukaryotic Gene Architecture
Abstract
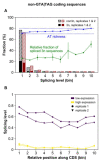
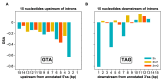
The conventionally clear distinction between exons and introns in eukaryotic genes is actually blurred. To illustrate this point, consider sequences that are retained in mature mRNAs about 50% of the time: how should they be classified? Moreover, although it is clear that RNA splicing influences gene expression levels and is an integral part of interdependent cellular networks, introns continue to be regarded as accidental insertions; exogenous sequences whose evolutionary origin is independent of mRNA-associated processes and somewhat still elusive. Here, we present evidence that aids to resolve this disconnect between conventional views about introns and current knowledge about the role of RNA splicing in the eukaryotic cell. We first show that coding sequences flanked by cryptic splice sites are negatively selected on a genome-wide scale in Paramecium. Then, we exploit selection intensity to infer splicing-related evolutionary dynamics. Our analyses suggest that intron gain begins as a splicing error, involves a transient phase of alternative splicing, and is preferentially completed at the 5' end of genes, which through intron gain can become highly expressed. We conclude that relaxed selective constraints may promote biological complexity in Paramecium and that the relationship between exons and introns is fluid on an evolutionary scale.
Keywords: RNA splicing; alternative splicing; exon; exonization; gene architecture; gene expression; intron; intronization; purifying selection.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
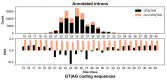
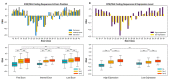
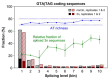
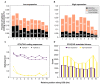
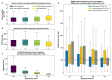
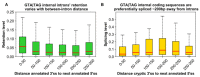
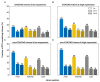
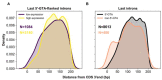
References
Grants and funding
LinkOut - more resources
Full Text Sources

