G-Quadruplex Aptamer-Ligand Characterization
- PMID: 36296374
- PMCID: PMC9609330
- DOI: 10.3390/molecules27206781
G-Quadruplex Aptamer-Ligand Characterization
Abstract
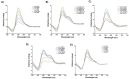
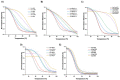
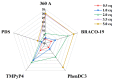
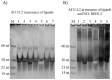
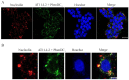
In this work we explore the structure of a G-rich DNA aptamer termed AT11-L2 (TGGTGGTGGTTGTTGTTGGTGGTGGTGGT; derivative of AT11) by evaluating the formation and stability of G-quadruplex (G4) conformation under different experimental conditions such as KCl concentration, temperature, and upon binding with a variety of G4 ligands (360A, BRACO-19, PDS, PhenDC3, TMPyP4). We also determined whether nucleolin (NCL) can be a target of AT11-L2 G4. Firstly, we assessed by circular dichroism, UV and NMR spectroscopies the formation of G4 by AT11-L2. We observed that, for KCl concentrations of 65 mM or less, AT11-L2 adopts hybrid or multiple topologies. In contrast, a parallel topology predominates for buffer containing 100 mM of KCl. The Tm of AT11-L2 in 100 mM of KCl is 38.9 °C, proving the weak stability of this sequence. We also found that upon titration with two molar equivalents of 360A, BRACO-19 and PhenDC3, the G4 is strongly stabilized and its topology is maintained, while the addition of 3.5 molar equivalents of TMPyP4 promotes the disruption of G4. The KD values between AT11-L2 G4, ligands and NCL were obtained by fluorescence titrations and are in the range of µM for ligand complexes and nM when adding NCL. In silico studies suggest that four ligands bind to the AT11-L2 G4 structure by stacking interactions, while the RBD1,2 domains of NCL interact preferentially with the thymines of AT11-L2 G4. Finally, AT11-L2 G4 co-localized with NCL in NCL-positive tongue squamous cell carcinoma cell line.
Keywords: G-quadruplex aptamer; aptamer–ligand interactions; biophysical techniques; ligands.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

