Preprocessing NIR Spectra for Aquaphotomics
- PMID: 36296387
- PMCID: PMC9610546
- DOI: 10.3390/molecules27206795
Preprocessing NIR Spectra for Aquaphotomics
Abstract
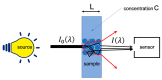
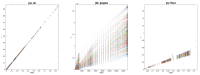
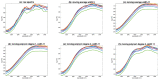
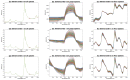
Even though NIR spectroscopy is based on the Beer-Lambert law, which clearly relates the concentration of the absorbing elements with the absorbance, the measured spectra are subject to spurious signals, such as additive and multiplicative effects. The use of NIR spectra, therefore, requires a preprocessing step. This article reviews the main preprocessing methods in the light of aquaphotomics. Simple methods for visualizing the spectra are proposed in order to guide the user in the choice of the best preprocessing. The most common chemometrics preprocessing are presented and illustrated by three real datasets. Some preprocessing aims to produce a spectrum as close as possible to the absorbance that would have been measured under ideal conditions and is very useful for the establishment of an aquagram. Others, dedicated to the improvement of the resolution of the spectra, are very useful for the identification of the peaks. Finally, special attention is given to the problem of reducing multiplicative effects and to the potential pitfalls of some very popular methods in chemometrics. Alternatives proposed in recent papers are presented.
Keywords: aquaphotomics; chemometrics; near infrared spectroscopy; preprocessing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






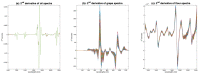
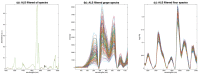

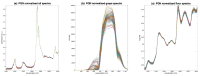
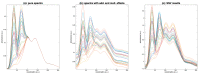

References
-
- Siesler H.W., Kawata S., Heise H.M., Ozaki Y., editors. Near-Infrared Spectroscopy: Principles, Instruments, Applications. John Wiley & Sons; Hoboken, NJ, USA: 2008.
-
- Tsenkova R. Visible-near infrared perturbation spectroscopy: Water in action seen as a source of information; Proceedings of the 12th International Conference on Near-Infrared Spectroscopy; Auckland, New Zealand. 1 September 2005; pp. 607–612.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

