Genome-Wide Identification and Expression Analysis of HSF Transcription Factors in Alfalfa (Medicago sativa) under Abiotic Stress
- PMID: 36297789
- PMCID: PMC9609925
- DOI: 10.3390/plants11202763
Genome-Wide Identification and Expression Analysis of HSF Transcription Factors in Alfalfa (Medicago sativa) under Abiotic Stress
Abstract
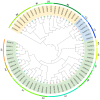
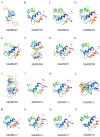
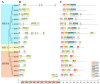
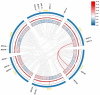
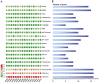
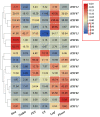
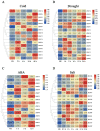
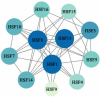
Alfalfa (Medicago sativa) is one of the most important legume forage species in the world. It is often affected by several abiotic stressors that result in reduced yields and poor growth. Therefore, it is crucial to study the resistance of M. sativa to abiotic stresses. Heat shock transcription factors (HSF) are key players in a number of transcriptional regulatory pathways. These pathways play an essential role in controlling how plants react to different abiotic stressors. Studies on the HSF gene family have been reported in many species but have not yet undergone a thorough analysis in M. sativa. Therefore, in order to identify a more comprehensive set of HSF genes, from the genomic data, we identified 16 members of the MsHSF gene, which were unevenly distributed over six chromosomes. We also looked at their gene architectures and protein motifs, and phylogenetic analysis allowed us to divide them into 3 groups with a total of 15 subgroups. Along with these aspects, we then examined the physicochemical properties, subcellular localization, synteny analysis, GO annotation and enrichment, and protein interaction networks of amino acids. Finally, the analysis of 16 MsHSF genes' expression levels across all tissues and under four abiotic stresses using publicly available RNA-Seq data revealed that these genes had significant tissue-specific expression. Moreover, the expression of most MsHSF genes increased dramatically under abiotic stress, further validating the critical function played by the MsHSF gene family in abiotic stress. These results provided basic information about MsHSF gene family and laid a foundation for further study on the biological role of MsHSF gene in response to stress in M. sativa.
Keywords: HSF gene family; Medicago sativa; abiotic stress; expression profile.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Song Y., Lv J., Ma Z., Dong W. The mechanism of alfalfa (Medicago sativa L.) response to abiotic stress. Plant Growth Regul. 2019;89:239–249. doi: 10.1007/s10725-019-00530-1. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

