Characterization and Pathogenicity of Two Novel PRRSVs Recombined by NADC30-like and NADC34-like Strains in China
- PMID: 36298730
- PMCID: PMC9607012
- DOI: 10.3390/v14102174
Characterization and Pathogenicity of Two Novel PRRSVs Recombined by NADC30-like and NADC34-like Strains in China
Abstract
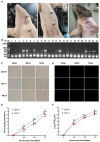
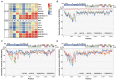
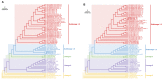
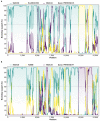
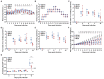
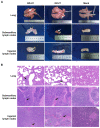
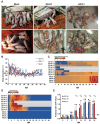
Porcine reproductive and respiratory syndrome viruses (PRRSVs) pose a serious threat to the swine industry in China, which has caused great difficulties for porcine reproductive and respiratory syndrome (PRRS) immune prevention and control, due to its easily mutable and recombinant nature. In this study, two novel PRRSV strains, which were named GD-H1 and GD-F1, were isolated and fully sequenced from pig farms in Guangdong province, China. The phylogenetic analysis and recombination analysis revealed that the GD-H1 and GD-F1 were generated by the recombination of NADC30-like and NADC34-like strains which were different from the previously prevalent strain. Further pathogenic studies on piglets and sows found that the recombinant strains could cause piglets high fever, loss of appetite and lung lesions, but no piglets died. However, the recombinant strains could cause acute death and abortion in pregnant sow infection models together with average survival rates of 62.5% and 37.5% abortion rates, respectively. These findings indicated that the recombinant strains were extremely pathogenic to sows. Therefore, we report two clinical novel recombinant strains of PRRSV that are different from the traditional epidemic strains in China, which may provide early warning and support for PRRS immune prevention and control.
Keywords: PRRSV; pathogenicity; phylogenetic analyses; recombination analyses.
Conflict of interest statement
The authors declare no potential conflict of interest with respect to the research, authorship and publication of this article.
Figures

References
-
- Lole K.S., Bollinger R.C., Paranjape R.S., Gadkari D., Kulkarni S.S., Novak N.G., Ingersoll R., Sheppard H.W., Ray S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with Evidence of Intersubtype Recombination. J. Virol. 1999;73:152–160. doi: 10.1128/JVI.73.1.152-160.1999. - DOI - PMC - PubMed
-
- Gao J.-C., Xiong J.-Y., Ye C., Chang X.-B., Guo J.-C., Jiang C.-G., Zhang G.-H., Tian Z.-J., Cai X.-H., Tong G.-Z., et al. Genotypic and geographical distribution of porcine reproductive and respiratory syndrome viruses in mainland China in 1996–2016. Vet. Microbiol. 2017;208:164–172. doi: 10.1016/j.vetmic.2017.08.003. - DOI - PubMed
-
- Firth A.E., Zevenhoven-Dobbe J.C., Wills N.M., Go Y.Y., Balasuriya U.B.R., Atkins J., Snijder E.J., Posthuma C.C. Discovery of a small arterivirus gene that overlaps the GP5 coding sequence and is important for virus production. J. Gen. Virol. 2011;92:1097–1106. doi: 10.1099/vir.0.029264-0. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

