Phylodynamics of Highly Pathogenic Avian Influenza A(H5N1) Virus Circulating in Indonesian Poultry
- PMID: 36298771
- PMCID: PMC9608721
- DOI: 10.3390/v14102216
Phylodynamics of Highly Pathogenic Avian Influenza A(H5N1) Virus Circulating in Indonesian Poultry
Abstract
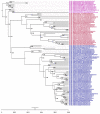
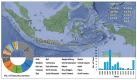
After its first detection in 1996, the highly pathogenic avian influenza A(H5Nx) virus has spread extensively worldwide. HPAIv A(H5N1) was first detected in Indonesia in 2003 and has been endemic in poultry in this country ever since. However, Indonesia has limited information related to the phylodynamics of HPAIv A(H5N1) in poultry. The present study aimed to increase the understanding of the evolution and temporal dynamics of HPAIv H5N1 in Indonesian poultry between 2003 and 2016. To this end, HPAIv A(H5N1) hemagglutinin sequences of viruses collected from 2003 to 2016 were analyzed using Bayesian evolutionary analysis sampling trees. Results indicated that the common ancestor of Indonesian poultry HPAIv H5N1 arose approximately five years after the common ancestor worldwide of HPAI A(H5Nx). In addition, this study indicated that only two introductions of HPAIv A(H5N1) occurred, after which these viruses continued to evolve due to extensive spread among poultry. Furthermore, this study revealed the divergence of H5N1 clade 2.3.2.1c from H5N1 clade 2.3.2.1b. Both clades 2.3.2.1c and 2.3.2.1b share a common ancestor, clade 1, suggesting that clade 2.3.2.1 originated and diverged from China and other Asian countries. Since there was limited sequence and surveillance data for the HPAIv A(H5N1) from wild birds in Indonesia, the exact role of wild birds in the spread of HPAIv in Indonesia is currently unknown. The evolutionary dynamics of the Indonesian HPAIv A(H5N1) highlight the importance of continuing and improved genomic surveillance and adequate control measures in the different regions of both the poultry and wild birds. Spatial genomic surveillance is useful to take adequate control measures. Therefore, it will help to prevent the future evolution of HPAI A(H5N1) and pandemic threats.
Keywords: Bayesian evolutionary analysis; H5N1; HPAI; Indonesia; phylodynamic.
Conflict of interest statement
The authors declare that the research was conducted in the absence of potential conflicts of interest.
Figures


References
-
- WHO . Cumulative Number of Confirmed Human Cases for Avian Influenza A(H5N1) Reported to WHO, 2003–2022. WHO; Geneva, Switzerland: 2022.
-
- Wibawa H., Karo-Karo D., Pribadi E.S., Bouma A., Bodewes R., Vernooij H., Sugama A., Muljono D.H., Koch G. Exploring contacts facilitating transmission of influenza A(H5N1) virus between poultry farms in West Java, Indonesia: A major role for backyard farms? Prev. Veter.-Med. 2018;156:8–15. doi: 10.1016/j.prevetmed.2018.04.008. - DOI - PubMed
-
- Karo-Karo D., Diyantoro, Pribadi E.S., Sudirman F.X., Kurniasih S.W., Sukirman, Indasari I., Muljono D.H., Koch G., Stegeman J.A. Highly Pathogenic Avian Influenza A(H5N1) Outbreaks in West Java Indonesia 2015–2016: Clinical Manifestation and Associated Risk Factors. Microorganisms. 2019;7:327. doi: 10.3390/microorganisms7090327. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

