Characterization of Grapevine Fanleaf Virus Isolates in 'Chardonnay' Vines Exhibiting Severe and Mild Symptoms in Two Vineyards
- PMID: 36298857
- PMCID: PMC9609649
- DOI: 10.3390/v14102303
Characterization of Grapevine Fanleaf Virus Isolates in 'Chardonnay' Vines Exhibiting Severe and Mild Symptoms in Two Vineyards
Abstract
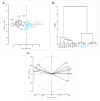
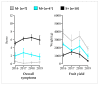
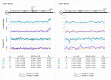
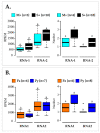
Fanleaf degeneration is a complex viral disease of Vitis spp. that detrimentally impacts fruit yield and reduces the productive lifespan of most vineyards worldwide. In France, its main causal agent is grapevine fanleaf virus (GFLV). In the past, field experiments were conducted to explore cross-protection as a management strategy of fanleaf degeneration, but results were unsatisfactory because the mild virus strain negatively impacted fruit yield. In order to select new mild GFLV isolates, we examined two old 'Chardonnay' parcels harbouring vines with distinct phenotypes. Symptoms and agronomic performances were monitored over the four-year study on 21 individual vines that were classified into three categories: asymptomatic GFLV-free vines, GFLV-infected vines severely diseased and GFLV-infected vines displaying mild symptoms. The complete coding genomic sequences of GFLV isolates in infected vines was determined by high-throughput sequencing. Most grapevines were infected with multiple genetically divergent variants. While no specific molecular features were apparent for GFLV isolates from vines displaying mild symptoms, a genetic differentiation of GFLV populations depending on the vineyard parcel was observed. The mild symptomatic grapevines identified during this study were established in a greenhouse to recover GFLV variants of potential interest for cross-protection studies.
Keywords: cross-protection; fanleaf degeneration; genetic and phenotypic diversity; grapevine; grapevine fanleaf virus; high throughput sequencing; mild isolates; symptomatology; virome.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Martelli G.P. Virus Diseases of Grapevine. John Wiley & Sons, Ltd.; Hoboken, NJ, USA: 2014. - DOI
-
- Martelli G.P. An overview on grapevine viruses, viroids, and the diseases they cause. In: Meng B., Martelli G.P., Golino D.A., Fuchs M., editors. Grapevine Viruses: Molecular Biology, Diagnostics and Management. Springer International Publishing; Cham, Switzerland: 2017. pp. 31–46.
-
- Fuchs M. Grapevine viruses: A multitude of diverse species with simple but overall poorly adopted management solutions in the vineyard. J. Plant Pathol. 2020;102:643–653. doi: 10.1007/s42161-020-00579-2. - DOI
-
- Demangeat G., Esmenjaud D., Voisin R., Bidault J.-M., Grenan S., Claverie M. Le court-noué de la vigne: État des connaissances sur cette maladie. Phytoma. 2005;587:38–42.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

