Impact of Early Pandemic SARS-CoV-2 Lineages Replacement with the Variant of Concern P.1 (Gamma) in Western Bahia, Brazil
- PMID: 36298869
- PMCID: PMC9611628
- DOI: 10.3390/v14102314
Impact of Early Pandemic SARS-CoV-2 Lineages Replacement with the Variant of Concern P.1 (Gamma) in Western Bahia, Brazil
Abstract
Background: The correct understanding of the epidemiological dynamics of COVID-19, caused by the SARS-CoV-2, is essential for formulating public policies of disease containment.
Methods: In this study, we constructed a picture of the epidemiological dynamics of COVID-19 in a Brazilian population of almost 17000 patients in 15 months. We specifically studied the fluctuations of COVID-19 cases and deaths due to COVID-19 over time according to host gender, age, viral load, and genetic variants.
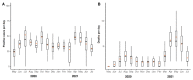
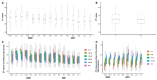
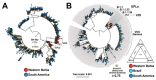
Results: As the main results, we observed that the numbers of COVID-19 cases and deaths due to COVID-19 fluctuated over time and that men were the most affected by deaths, as well as those of 60 or more years old. We also observed that individuals between 30- and 44-years old were the most affected by COVID-19 cases. In addition, the viral loads in the patients' nasopharynx were higher in the early symptomatic period. We found that early pandemic SARS-CoV-2 lineages were replaced by the variant of concern (VOC) P.1 (Gamma) in the second half of the study period, which led to a significant increase in the number of deaths.
Conclusions: The results presented in this study are helpful for future formulations of efficient public policies of COVID-19 containment.
Keywords: COVID-19; impact; variant of concern.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B., et al. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. - PMC - PubMed
-
- WHO Coronavirus (COVID-19) Dashboard|WHO Coronavirus Disease (COVID-19) Dashboard. [(accessed on 26 September 2022)]. Available online: https://covid19.who.int/
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

