Construction of an immune-related signature for predicting the ischemic events in patients undergoing carotid endarterectomy
- PMID: 36299596
- PMCID: PMC9592116
- DOI: 10.3389/fgene.2022.1014264
Construction of an immune-related signature for predicting the ischemic events in patients undergoing carotid endarterectomy
Erratum in
-
Corrigendum: Construction of an immune-related signature for predicting the ischemic events in patients undergoing carotid endarterectomy.Front Genet. 2023 Jan 6;13:1104846. doi: 10.3389/fgene.2022.1104846. eCollection 2022. Front Genet. 2023. PMID: 36685906 Free PMC article.
Abstract
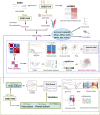
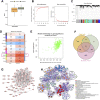
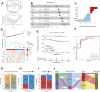
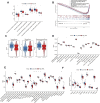
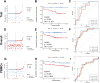
Background: Inflammatory responses have drawn more attention to atherosclerosis; however, the immune-related genes (IRGs) as a prognostic factor in atherosclerotic plaque remain to be fully elucidated. Here, the purpose of this study was to investigate whether the IRGs could be identified as a reliable biomarker for predicting ischemic events in patients undergoing carotid endarterectomy (CEA). Methods: Two datasets GSE97210 and GSE21545 were downloaded from the Gene Expression Omnibus (GEO) database. The dataset GSE97210 was used to explore the significant pathways and differentially expressed IRGs (DEIRGs) between plaques and controls, which were further screened to identify the prognostic DEIRGs in the GSE21545 dataset. The identification of molecular subgroups with the prognostic gene expression patterns was achieved through nonnegative matrix factorization (NMF) clustering. Functional analyses including GO, KEGG, GSVA, and GSEA analyses, and immune analyses including xCell and ssGSEA algorithms were conducted to elucidate the underlying mechanisms. The prognostic risk model was constructed using the LASSO algorithm and multivariate Cox regression analysis. Results: A total of 796 DEIRGs (including 588 upregulated and 208 downregulated) were identified. Nine prognostic DEIRGs were further screened with univariate Cox regression analysis. Two clusters with different prognosis were grouped based on the prognostic DEIRGs. Immune infiltration analysis shows that cluster 2 with a better prognosis presented with a higher immune response than cluster 1. A prognostic model based on seven IRGs (IL2RA, NR4A2, DES, ERAP2, SLPI, RASGRP1, and AGTR2) was developed and verified. Consistent with the immune analysis of the cluster, the immune infiltration in the low-risk group with a better prognosis was also more active than that in the high-risk group. Finally, a nomogram based on the seven genes was constructed, which might have future implications in clinical care. Conclusion: The expression of immune-related genes is correlated with the immune microenvironment of atherosclerotic patients and could be applied to predict the ischemic events in patients undergoing CEA accurately.
Keywords: atherosclerosis; immune infiltration; immune-related genes; ischemic events; molecular subtypes.
Copyright © 2022 Li, Zhang, Weng and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Bai H. L., Lu Z. F., Zhao J. J., Ma X., Li X. H., Xu H., et al. (2019). Microarray profiling analysis and validation of novel long noncoding RNAs and mRNAs as potential biomarkers and their functions in atherosclerosis. Physiol. Genomics 51, 644–656. 10.1152/physiolgenomics.00077.2019 - DOI - PMC - PubMed
-
- Bakhshi H., Varadarajan V., Ambale-Venkatesh B., Meyghani Z., Ostovaneh M. R., Durda P., et al. (2020). Association of soluble interleukin-2 receptor α and tumour necrosis factor receptor 1 with heart failure: The Multi-Ethnic Study of Atherosclerosis. Esc. Heart Fail. 7, 639–644. 10.1002/ehf2.12623 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

