Transcriptome analysis of malate-induced Schizochytrium sp. FJU-512 reveals a novel pathway for biosynthesis of docosahexaenoic acid with enhanced expression of genes responsible for acetyl-CoA and NADPH accumulation
- PMID: 36299719
- PMCID: PMC9589357
- DOI: 10.3389/fmicb.2022.1006138
Transcriptome analysis of malate-induced Schizochytrium sp. FJU-512 reveals a novel pathway for biosynthesis of docosahexaenoic acid with enhanced expression of genes responsible for acetyl-CoA and NADPH accumulation
Abstract
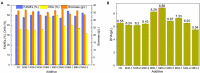
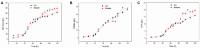
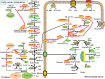
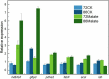
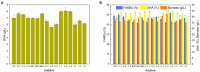
Schizochytrium is one of the few oleaginous microalgae that produce docosahexaenoic acid (DHA)-rich lipids. In this study, global changes in gene expression levels of Schizochytrium sp. FJU-512 cultured with malate in a 15 l-bioreactor was analyzed using comparative transcriptomics. The changes were found mainly in the genes involved in oxidative phosphorylation, β-oxidation, and pentose phosphate pathways. Consequently, the global changes in genes associated with the pathways could lead to an increase in the influx throughputs of pyruvate, branched-chain amino acids, fatty acids, and vitamin B6. Our transcriptome analysis indicated pyruvate dehydrogenase E2 component and acetolactate synthase I/II/III large subunit as major contributors to acetyl-CoA biosynthesis, whereas glucose-6-phosphate dehydrogenase was indicated as the major contributor to the biosynthesis of NADPH. An increase in DHA titer of up to 22% was achieved with the addition of malate to the fed-batch culture of Schizochytrium sp. FJU-512. This study provides an alternate method to enhance DHA production in Schizochytrium sp. FJU-512 through malate induced upregulation of genes responsible for acetyl-CoA and NADPH biosynthesis.
Keywords: acetyl-CoA; comparative transcriptomics; docosahexaenoic acid; fatty acid metabolism; malate.
Copyright © 2022 Zhang, Gao, Yu, Wang, Weng, Li, He, Guo, Zhang, Huang and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Metabolic engineering to enhance biosynthesis of both docosahexaenoic acid and odd-chain fatty acids in Schizochytrium sp. S31.Biotechnol Biofuels. 2019 Jun 8;12:141. doi: 10.1186/s13068-019-1484-x. eCollection 2019. Biotechnol Biofuels. 2019. PMID: 31182976 Free PMC article.
-
Transcriptome and gene expression analysis of docosahexaenoic acid producer Schizochytrium sp. under different oxygen supply conditions.Biotechnol Biofuels. 2018 Sep 17;11:249. doi: 10.1186/s13068-018-1250-5. eCollection 2018. Biotechnol Biofuels. 2018. PMID: 30245741 Free PMC article.
-
Low-temperature effects on docosahexaenoic acid biosynthesis in Schizochytrium sp. TIO01 and its proposed underlying mechanism.Biotechnol Biofuels. 2020 Oct 16;13:172. doi: 10.1186/s13068-020-01811-y. eCollection 2020. Biotechnol Biofuels. 2020. PMID: 33088342 Free PMC article.
-
The strategies to reduce cost and improve productivity in DHA production by Aurantiochytrium sp.: from biochemical to genetic respects.Appl Microbiol Biotechnol. 2020 Nov;104(22):9433-9447. doi: 10.1007/s00253-020-10927-y. Epub 2020 Sep 26. Appl Microbiol Biotechnol. 2020. PMID: 32978687 Review.
-
Advancing oleaginous microorganisms to produce lipid via metabolic engineering technology.Prog Lipid Res. 2013 Oct;52(4):395-408. doi: 10.1016/j.plipres.2013.05.002. Epub 2013 May 16. Prog Lipid Res. 2013. PMID: 23685199 Review.
Cited by
-
Metabolic regulation strategies for enhancing microbial docosahexaenoic acid production by Schizochytrium sp.World J Microbiol Biotechnol. 2025 Apr 28;41(5):142. doi: 10.1007/s11274-025-04268-z. World J Microbiol Biotechnol. 2025. PMID: 40289231 Review.
-
Thraustochytrids: Evolution, Ultrastructure, Biotechnology, and Modeling.Int J Mol Sci. 2024 Dec 7;25(23):13172. doi: 10.3390/ijms252313172. Int J Mol Sci. 2024. PMID: 39684882 Free PMC article. Review.
References
-
- Abbadi A., Brummel M., Schutt B. S., Slabaugh M. B., Schuch R., Spener F. (2000). Reaction mechanism of recombinant 3-oxoacyl-(acyl-carrier-protein) synthase III from Cuphea wrightii embryo, a fatty acid synthase type II condensing enzyme. Biochem. J. 345, 153–160. doi: 10.1042/bj3450153 - DOI - PMC - PubMed
-
- Bi Z. Q., Ren L. J., Hu X. C., Sun X. M., Zhu S. Y., Ji X. J., et al. . (2018). Transcriptome and gene expression analysis of docosahexaenoic acid producer Schizochytrium sp. under different oxygen supply conditions. Biotechnol. Biofuels 11:249. doi: 10.1186/s13068-018-1250-5, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

