Jingmenviruses: Ubiquitous, understudied, segmented flavi-like viruses
- PMID: 36299728
- PMCID: PMC9589506
- DOI: 10.3389/fmicb.2022.997058
Jingmenviruses: Ubiquitous, understudied, segmented flavi-like viruses
Abstract
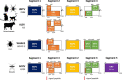
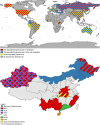
Jingmenviruses are a group of viruses identified recently, in 2014, and currently classified by the International Committee on Taxonomy of Viruses as unclassified Flaviviridae. These viruses closely related to flaviviruses are unique due to the segmented nature of their genome. The prototype jingmenvirus, Jingmen tick virus (JMTV), was discovered in Rhipicephalus microplus ticks collected from China in 2010. Jingmenviruses genomes are composed of four to five segments, encoding for up to seven structural proteins and two non-structural proteins, both of which display strong similarities with flaviviral non-structural proteins (NS2B/NS3 and NS5). Jingmenviruses are currently separated into two phylogenetic clades. One clade includes tick- and vertebrate-associated jingmenviruses, which have been detected in ticks and mosquitoes, as well as in humans, cattle, monkeys, bats, rodents, sheep, and tortoises. In addition to these molecular and serological detections, over a hundred human patients tested positive for jingmenviruses after developing febrile illness and flu-like symptoms in China and Serbia. The second phylogenetic clade includes insect-associated jingmenvirus sequences, which have been detected in a wide range of insect species, as well as in crustaceans, plants, and fungi. In addition to being found in various types of hosts, jingmenviruses are endemic, as they have been detected in a wide range of environments, all over the world. Taken together, all of these elements show that jingmenviruses correspond exactly to the definition of emerging viruses at risk of causing a pandemic, since they are already endemic, have a close association with arthropods, are found in animals in close contact with humans, and have caused sporadic cases of febrile illness in multiple patients. Despite these arguments, the vast majority of published data is from metagenomics studies and many aspects of jingmenvirus replication remain to be elucidated, such as their tropism, cycle of transmission, structure, and mechanisms of replication and restriction or epidemiology. It is therefore crucial to prioritize jingmenvirus research in the years to come, to be prepared for their emergence as human or veterinary pathogens.
Keywords: arbovirus; emerging virus; jingmenvirus; segmented flavivirus; tick-borne disease.
Copyright © 2022 Colmant, Charrel and Coutard.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

