Integrative genomic analysis facilitates precision strategies for glioblastoma treatment
- PMID: 36300002
- PMCID: PMC9589211
- DOI: 10.1016/j.isci.2022.105276
Integrative genomic analysis facilitates precision strategies for glioblastoma treatment
Abstract
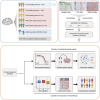
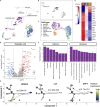
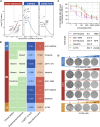
Glioblastoma (GBM) is the most common form of malignant primary brain tumor with a dismal prognosis. Currently, the standard treatments for GBM rarely achieve satisfactory results, which means that current treatments are not individualized and precise enough. In this study, a multiomics-based GBM classification was established and three subclasses (GPA, GPB, and GPC) were identified, which have different molecular features both in bulk samples and at single-cell resolution. A robust GBM poor prognostic signature (GPS) score model was then developed using machine learning method, manifesting an excellent ability to predict the survival of GBM. NVP-BEZ235, GDC-0980, dasatinib and XL765 were ultimately identified to have subclass-specific efficacy targeting patients with a high risk of poor prognosis. Furthermore, the GBM classification and GPS score model could be considered as potential biomarkers for immunotherapy response. In summary, an integrative genomic analysis was conducted to advance individual-based therapies in GBM.
Keywords: Artificial intelligence; Cancer; Cancer systems biology; Omics.
© 2022 The Authors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Abdelfattah N., Kumar P., Wang C., Leu J.S., Flynn W.F., Gao R., Baskin D.S., Pichumani K., Ijare O.B., Wood S.L., et al. Single-cell analysis of human glioma and immune cells identifies S100A4 as an immunotherapy target. Nat. Commun. 2022;13:767. doi: 10.1038/s41467-022-28372-y. - DOI - PMC - PubMed
-
- Alhalabi O.T., Fletcher M.N.C., Hielscher T., Kessler T., Lokumcu T., Baumgartner U., Wittmann E., Schlue S., Göttmann M., Rahman S., et al. A novel patient stratification strategy to enhance the therapeutic efficacy of dasatinib in glioblastoma. Neuro Oncol. 2021;24:39–51. doi: 10.1093/neuonc/noab158. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

