A novel NF2 splicing mutant causes neurofibromatosis type 2 via liquid-liquid phase separation with large tumor suppressor and Hippo pathway
- PMID: 36300003
- PMCID: PMC9589172
- DOI: 10.1016/j.isci.2022.105275
A novel NF2 splicing mutant causes neurofibromatosis type 2 via liquid-liquid phase separation with large tumor suppressor and Hippo pathway
Abstract
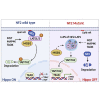
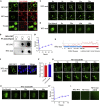
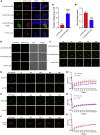
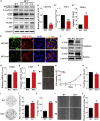
Neurofibromatosis type 2 is an autosomal dominant multiple neoplasia syndrome and is usually caused by mutations in the neurofibromin 2 (NF2) gene, which encodes a tumor suppressor and initiates the Hippo pathway. However, the mechanism by which NF2 functions in the Hippo pathway isn't fully understood. Here we identified a NF2 c.770-784del mutation from a neurofibromatosis type 2 family. MD simulations showed that this mutation significantly changed the structure of the F3 module of the NF2-FERM domain. Functional assays indicated that the NF2 c.770-784del variant formed LLPS in the cytoplasm with LATS to restrain LATS plasma membrane localization and inactivated the Hippo pathway. Besides, this deletion partly caused a skipping of exon 8 and reduced the protein level of NF2, collectively promoting proliferation and tumorigenesis of meningeal cells. We identified an unrecognized mechanism of LLPS and splicing skipping for the NF2-induced Hippo pathway, which provided new insight into the pathogenesis of neurofibromatosis type 2.
Keywords: Clinical genetics; Functional aspects of cell biology; Pathophysiology.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Conformational flexibility determines the Nf2/merlin tumor suppressor functions.Matrix Biol Plus. 2021 Jul 1;12:100074. doi: 10.1016/j.mbplus.2021.100074. eCollection 2021 Dec. Matrix Biol Plus. 2021. PMID: 34337379 Free PMC article.
-
Prediction and evaluation of deleterious and disease causing non-synonymous SNPs (nsSNPs) in human NF2 gene responsible for neurofibromatosis type 2 (NF2).J Biomol Struct Dyn. 2021 Nov;39(18):7044-7055. doi: 10.1080/07391102.2020.1805018. Epub 2020 Aug 13. J Biomol Struct Dyn. 2021. PMID: 32787631
-
A deletion causing NF2 exon 9 skipping is associated with familial autosomal dominant intramedullary ependymoma.Neuro Oncol. 2014 Jan;16(2):250-5. doi: 10.1093/neuonc/not165. Epub 2013 Dec 18. Neuro Oncol. 2014. PMID: 24357459 Free PMC article.
-
Neurofibromatosis Type 2 (NF2) and the Implications for Vestibular Schwannoma and Meningioma Pathogenesis.Int J Mol Sci. 2021 Jan 12;22(2):690. doi: 10.3390/ijms22020690. Int J Mol Sci. 2021. PMID: 33445724 Free PMC article. Review.
-
Molecular insights into NF2/Merlin tumor suppressor function.FEBS Lett. 2014 Aug 19;588(16):2743-52. doi: 10.1016/j.febslet.2014.04.001. Epub 2014 Apr 12. FEBS Lett. 2014. PMID: 24726726 Free PMC article. Review.
Cited by
-
Emerging regulatory mechanisms and functions of biomolecular condensates: implications for therapeutic targets.Signal Transduct Target Ther. 2025 Jan 6;10(1):4. doi: 10.1038/s41392-024-02070-1. Signal Transduct Target Ther. 2025. PMID: 39757214 Free PMC article. Review.
-
High de novo mutation rate in Iranian NF2-related schwannomatosis patients with a report of a novel NF2 mutation.Mol Biol Rep. 2025 Apr 18;52(1):402. doi: 10.1007/s11033-025-10431-4. Mol Biol Rep. 2025. PMID: 40249415
-
New Insights into YAP/TAZ-TEAD-Mediated Gene Regulation and Biological Processes in Cancer.Cancers (Basel). 2023 Nov 21;15(23):5497. doi: 10.3390/cancers15235497. Cancers (Basel). 2023. PMID: 38067201 Free PMC article. Review.
-
Liquid-liquid phase separation: an emerging perspective on the tumorigenesis, progression, and treatment of tumors.Front Immunol. 2025 Jun 26;16:1604015. doi: 10.3389/fimmu.2025.1604015. eCollection 2025. Front Immunol. 2025. PMID: 40642070 Free PMC article. Review.
-
NF2 can mediate the expression of CAMK2A in a tissue specific manner.Sci Rep. 2024 Dec 30;14(1):31992. doi: 10.1038/s41598-024-83553-7. Sci Rep. 2024. PMID: 39738767 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

