Identification of a dysregulated CircRNA-associated gene signature for predicting prognosis, immune landscape, and drug candidates in bladder cancer
- PMID: 36300085
- PMCID: PMC9589509
- DOI: 10.3389/fonc.2022.1018285
Identification of a dysregulated CircRNA-associated gene signature for predicting prognosis, immune landscape, and drug candidates in bladder cancer
Abstract
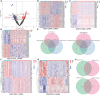
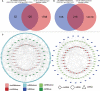
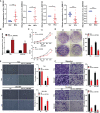
Increasing evidences have demonstrated that circular RNA (circRNAs) plays a an essential regulatory role in initiation, progression and immunotherapy resistance of various cancers. However, circRNAs have rarely been studied in bladder cancer (BCa). The purpose of this research is to explore new circRNAs and their potential mechanisms in BCa. A novel ceRNA-regulated network, including 87 differentially expressed circRNAs (DE-circRNAs), 126 DE-miRNAs, and 217 DE-mRNAs was constructed to better understanding the biological processes using Cytoscape 3.7.1 based on our previously high-throughput circRNA sequencing and five GEO datasets. Subsequently, five randomly selected circRNAs (upregulated circ_0001681; downregulated circ_0000643, circ_0001798, circ_0006117 and circ_0067900) in 20 pairs of BCa and paracancerous tissues were confirmed using qRT-PCR. Functional analysis results determined that 772 GO functions and 32 KEGG pathways were enriched in the ceRNA network. Ten genes (PFKFB4, EDNRA, GSN, GAS1, PAPPA, DTL, TGFBI, PRSS8, RGS1 and TCF4) were selected for signature construction among the ceRNA network. The Human Protein Atlas (HPA) expression of these genes were consistent with the above sequencing data. Notably, the model was validated in multiple external datasets (GSE13507, GSE31684, GSE48075, IMvigor210 and GSE32894). The immune-infiltration was evaluated by 7 published algorithms (i.e., TIMER, CIBERSORT, CIBERSORT-ABS, QUANTISEQ, MCPCOUNTER, XCELL and EPIC). Next, Correlations between riskscore or risk groups and clinicopathological data, overall survival, recognized immunoregulatory cells or common chemotherapeutic agents of BCa patients were performed using wilcox rank test, chi-square test, cox regression and spearman's correlation analysis; and, these results are significant. According to R package "GSVA" and "clusterProfiler", the most significantly enriched HALLMARK and KEGG pathway was separately the 'Epithelial Mesenchymal Transition' and 'Ecm Receptor Interaction' in the high- vs. low-risk group. Additionally, the functional experiments in vitro also revealed that the overexpression of has_circ_0067900 significantly impaired the proliferation, migration, and invasion capacities of BCa cells. Collectively, the results of the current study provide a novel landscape of circRNA-associated ceRNA-regulated network in BCa. The ceRNA-associated gene model which was constructed presented a high predictive performance for the prognosis, immunotherapeutic responsiveness, and chemotherapeutic sensitivity of BCa. And, has_circ_0067900 was originally proposed as tumor suppressor for patients with BCa.
Keywords: CircRNA-related prognostic signature; bladder cancer; chemotherapy; immune infiltration; immunotherapy.
Copyright © 2022 Shen, Li, Zhang, Zhang, Wu, Da, Yang, Wang, Zhang, Qie, Zhao, Lin, Huang, Zhou and Hu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







Similar articles
-
Development and validation of a glycolysis-associated gene signature for predicting the prognosis, immune landscape, and drug sensitivity in bladder cancer.Front Immunol. 2025 Jan 10;15:1430583. doi: 10.3389/fimmu.2024.1430583. eCollection 2024. Front Immunol. 2025. PMID: 39867879 Free PMC article.
-
Identification of Enzalutamide Resistance-Related circRNA-miRNA-mRNA Regulatory Networks in Patients with Prostate Cancer.Onco Targets Ther. 2021 Jun 21;14:3833-3848. doi: 10.2147/OTT.S309917. eCollection 2021. Onco Targets Ther. 2021. Retraction in: Onco Targets Ther. 2023 Jul 18;16:583-584. doi: 10.2147/OTT.S430430. PMID: 34188491 Free PMC article. Retracted.
-
Construction of circRNA-miRNA-mRNA network and identification of novel potential biomarkers for non-small cell lung cancer.Cancer Cell Int. 2021 Nov 20;21(1):611. doi: 10.1186/s12935-021-02278-z. Cancer Cell Int. 2021. PMID: 34801043 Free PMC article.
-
Expression profiles, biological functions and clinical significance of circRNAs in bladder cancer.Mol Cancer. 2021 Jan 4;20(1):4. doi: 10.1186/s12943-020-01300-8. Mol Cancer. 2021. PMID: 33397425 Free PMC article. Review.
-
CircRNA-Associated CeRNAs Regulatory Axes in Retinoblastoma: A Systematic Scoping Review.Front Oncol. 2022 Jul 5;12:910470. doi: 10.3389/fonc.2022.910470. eCollection 2022. Front Oncol. 2022. PMID: 35865469 Free PMC article.
Cited by
-
Non-coding RNAs in disease: from mechanisms to therapeutics.Nat Rev Genet. 2024 Mar;25(3):211-232. doi: 10.1038/s41576-023-00662-1. Epub 2023 Nov 15. Nat Rev Genet. 2024. PMID: 37968332 Review.
-
Circular RNA CCT3 is a unique molecular marker in bladder cancer.BMC Cancer. 2023 Oct 13;23(1):977. doi: 10.1186/s12885-023-11510-0. BMC Cancer. 2023. PMID: 37833621 Free PMC article.
-
Identifying stage-associated hub genes in bladder cancer via weighted gene co-expression network and robust rank aggregation analyses.Medicine (Baltimore). 2022 Dec 23;101(51):e32318. doi: 10.1097/MD.0000000000032318. Medicine (Baltimore). 2022. PMID: 36595851 Free PMC article.
-
CircRNAs in colorectal cancer: potential roles, clinical applications, and natural product-based regulation.Front Oncol. 2025 Jan 29;15:1525779. doi: 10.3389/fonc.2025.1525779. eCollection 2025. Front Oncol. 2025. PMID: 39944836 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

